Method for discriminating inversion carrying state of embryo chromosome
A chromosome and embryo technology, applied in the field of genetic diagnosis and human assisted reproduction, can solve the problem of not being able to accurately distinguish between embryos carrying chromosome inversions and completely normal embryos
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Example 1: Collection of Reference Samples from Patients and Parents of Patients
[0065] Two cases of chromosomal inversion carrier families who will undergo assisted reproduction were recruited, and the selected candidates were from the Obstetrics and Gynecology Hospital Affiliated to Fudan University and Shanghai Jiai Genetics and Infertility Diagnosis and Treatment Center. Each family was required to sign a written informed consent, and the research protocol was approved by the Human Subjects Ethics Committee of the Obstetrics and Gynecology Hospital Affiliated to Fudan University.
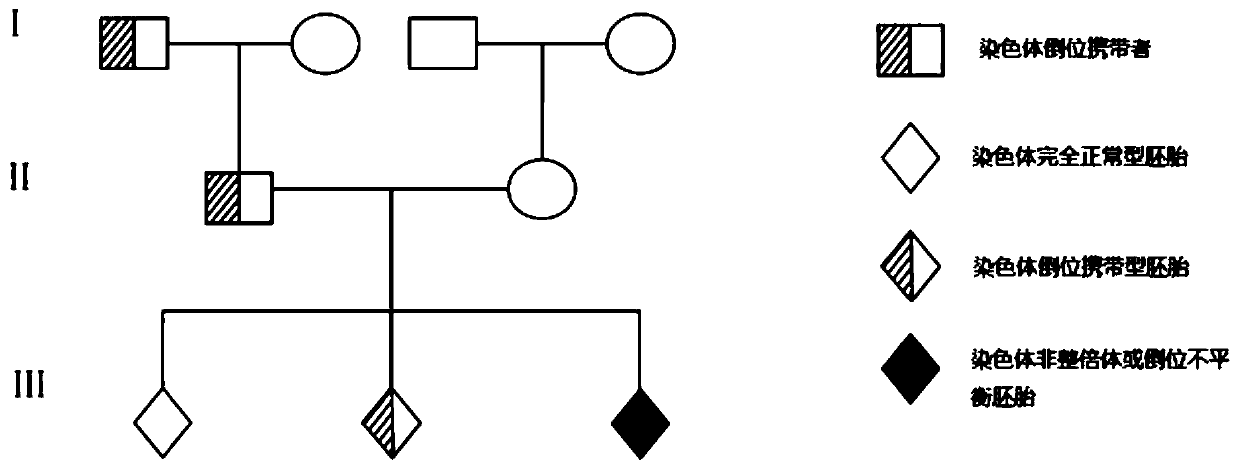
[0066] From June 2017 to June 2018, these two families had a history of recurrent spontaneous abortion or primary infertility. One of the couples carrying chromosomal inversion is referred to as "patient" and the other as "patient's spouse" in the following text , please refer to the family map carrying chromosomal inversion figure 1 . At the same time of recruitment, 10ml of peripher...
Embodiment 2
[0087] Example 2: Blastocyst Biopsy and Whole Genome Amplification (WGA)
[0088] 1. In vitro fertilization
[0089] In vitro fertilization (IVF) was carried out on the 2 recruited families, and the in vitro fertilization method followed the conventional methods in the field; the maternal / paternal age, phenotype, ovulation results, number of fertilized eggs and blastocysts finally used for biopsy in these families The quantities are listed in Table 2. Through the above in vitro fertilization, 2 families obtained a total of 6 blastocysts through multiple ovulation induction for subsequent biopsy and haplotype analysis.
[0090] Table 2. Basic information and in-vitro fertilization of families No. 1-2
[0091]
[0092] 2. Blastocyst biopsy and whole genome amplification
[0093] The above-mentioned embryos at the blastocyst stage were taken, and 3 to 10 cells were removed from the trophectoderm on the 5th or 6th day of embryonic development. Biopsied cells were placed in ...
Embodiment 3
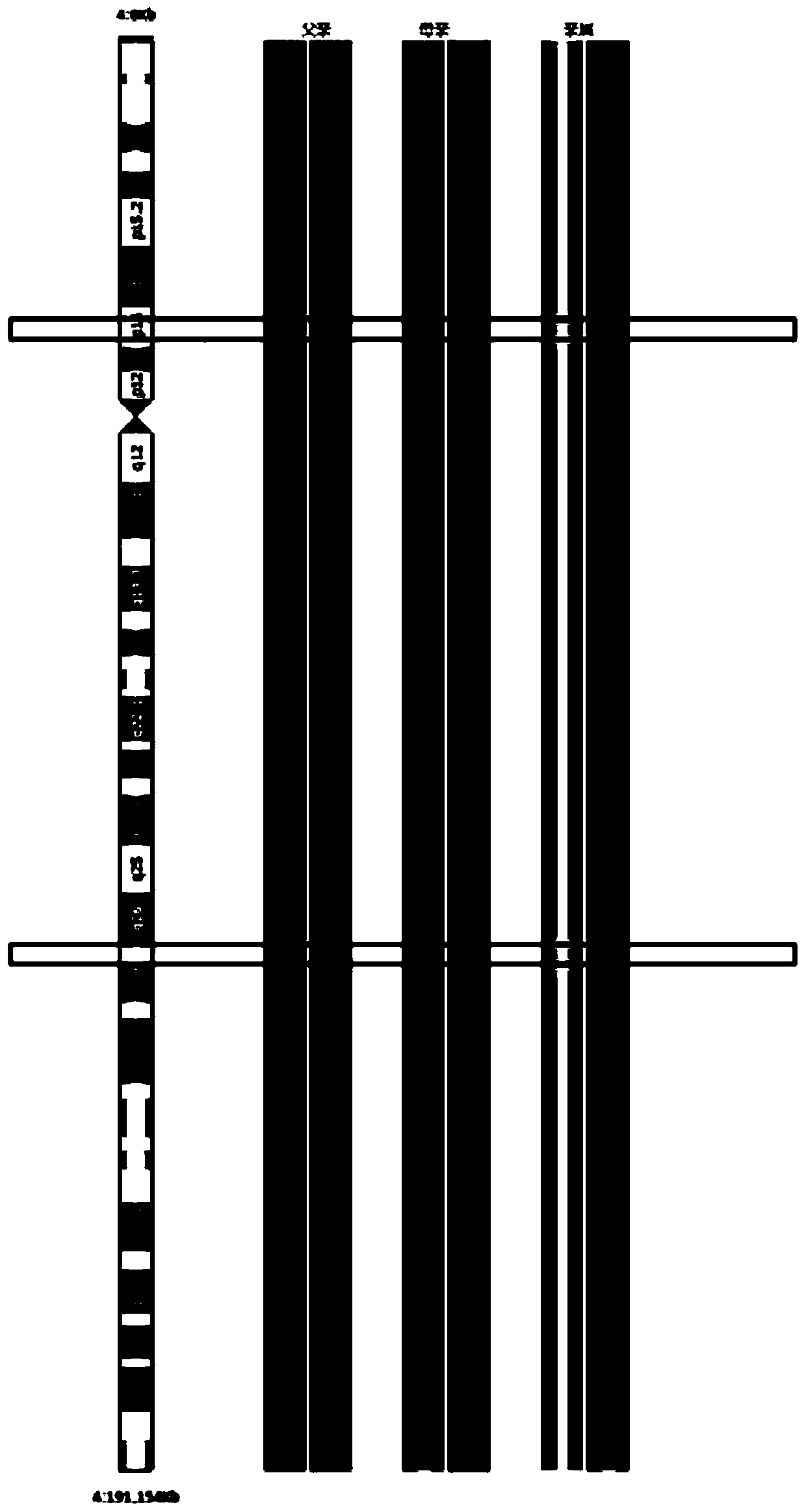
[0108] Example 3: SNP Genotyping and Haplotypes Analysis
[0109] 1. SNP genotype detection
[0110] SNP genotyping was performed using Illumina human Karyomap-12V1.0 microarray. Each Karyomap-12 chip contains nearly 300,000 SNPs, which can fully cover 23 pairs of human chromosomes. The 6 samples obtained from blastocyst biopsy and whole-genome amplification in Example 2 were grouped according to family, and were grouped with 2 whole-genome amplification samples of the patient couple and relatives of the family, respectively, for microarray SNP gene analysis. Type detection and analysis, the grouping situation is shown in Table 3. The specific experimental method is carried out with reference to the instructions, and will not be repeated here.
[0111] Table 3. SNP array experimental grouping of families 1-2
[0112]
[0113] Note: F stands for female, M stands for male
[0114] 2. Haplotype analysis
[0115] After obtaining all the SNPs information detected by the mi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com