A method for the detection of Pseudomonas aeruginosa based on aptamer fluorescence sensing
A Pseudomonas aeruginosa, fluorescence sensing technology, applied in fluorescence/phosphorescence, measuring device, material analysis by optical means, etc., can solve the problems of high cost, complex process, time-consuming, etc., and achieve the effect of easy production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
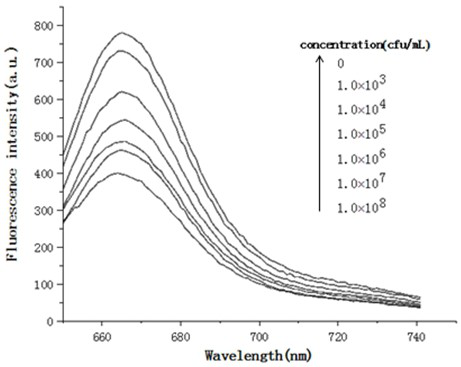
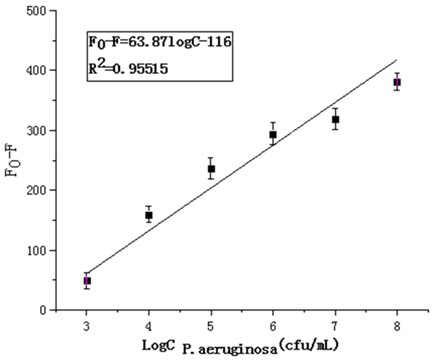
[0028] Embodiment 1 Real-time fluorescence detection of Pseudomonas aeruginosa live bacteria
[0029] A detection method for Pseudomonas aeruginosa, characterized in that it comprises the steps:
[0030] (1) Bacterial culture: resuscitate the purchased Pseudomonas aeruginosa freeze-dried powder, streak and inoculate them on the Lb nutrient agar plate and Lb liquid medium respectively, place the Lb nutrient agar plate in a refrigerator at 20°C for cultivation, and liquid culture The base was placed in a constant temperature incubator, set the temperature at 48°C, and the rotation speed was 150r / min, and cultivated for 48h. After shaking for a certain period of time, the culture is harvested and then used for detection. The obtained live bacterial suspension can be used directly or stored at 4°C;
[0031] (2) Count Pseudomonas aeruginosa colonies using the coating method, and calculate the bacterial concentration (CFU / mL) used for detection;
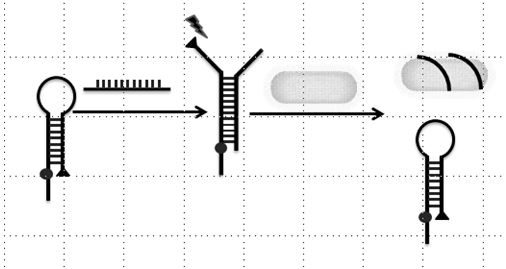
[0032] (3) The hairpin structure ...
Embodiment 2
[0037] Example 2 Real-time fluorescence detection of antibiotic inactivation of Pseudomonas aeruginosa
[0038] A detection method for antibiotic inactivation of Pseudomonas aeruginosa, characterized in that it comprises the steps:
[0039] (1) Bacterial culture: resuscitate the freeze-dried powder of Pseudomonas aeruginosa, streak and inoculate them on Lb nutrient agar plate and Lb liquid medium respectively, place the Lb nutrient agar plate in a refrigerator at 20°C for cultivation, and place the liquid medium in The constant temperature incubator is adjusted, the temperature is set at 48°C, the rotation speed is 150r / min, and the incubation is carried out for 48h. After shaking for a certain period of time, the cultures were harvested and used for assays. The resulting live bacterial suspension can be used directly or stored at 4°C;
[0040] (2) Count Pseudomonas aeruginosa colonies using the coating method, and the calculated colony concentration is 1.11*10 9 (CFU / mL); ...
Embodiment 3
[0047] Real-time fluorescence detection of Pseudomonas aeruginosa when embodiment 3 has interfering bacteria to exist
[0048] The detection method of Pseudomonas aeruginosa when interfering bacteria exists, is characterized in that comprising the steps:
[0049] (1) Bacterial culture: resuscitate the lyophilized powder of Pseudomonas aeruginosa, streak and inoculate them on Lb nutrient agar plate and Lb liquid medium respectively, place the Lb nutrient agar plate in a refrigerator at 20°C for cultivation, and inoculate Escherichia coli (BNCC336685 ) was recovered from the cryopreserved liquid, inoculated in Lb liquid medium, and the liquid medium was placed in a constant temperature incubator with a temperature of 48°C and a rotation speed of 150r / min, and cultured for 48 hours. After shaking for a certain period of time, the cultures were harvested and used for assays. The resulting live bacterial suspension can be used directly or stored at 4°C;
[0050] (2) Count the col...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com