Esterase gene and encoded protein as well as application
A technology of gene encoding and esterase, applied in the fields of recombinant plasmids, esterase genes, genetically engineered strains, and esterases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028]The cloning of embodiment 1 esterase EST-T2 gene, the construction of expression vector
[0029] (a) According to the genome sequence of the Bacillus subtilis endophyte HB-T2, clone the degrading PAE esterase gene EST-T2, the nucleotide sequence of which is shown in Seq ID NO.1.
[0030] The sequences of the upstream and downstream primers are respectively shown in SEQ ID NO.3 and SEQ ID NO.4, and the two primers respectively have BamHI and HindIII restriction sites to amplify the complete sequence of the EST-T2 gene.
[0031] Amplification system: (Beijing Qingke Xinye Biotechnology Co., Ltd. kit (TSE003):: 2×TSINGKE TM MasterMix25ul; 10uMprimerF / R1ul; DNA2 Oupto50ul); Amplification program: pre-denaturation at 94°C for 5min; cycle parameters: denaturation at 94°C for 30s, annealing at 57°C for 30s, extension at 72°C for 90s; run for 30 cycles; extension at 72°C for 10min, stabilization at 4°C for 1min, and the reaction ended.
[0032] The purified PCR product (Beijing...
Embodiment 2
[0037] Induced expression and purification of embodiment 2 esterase EST-T2 protein
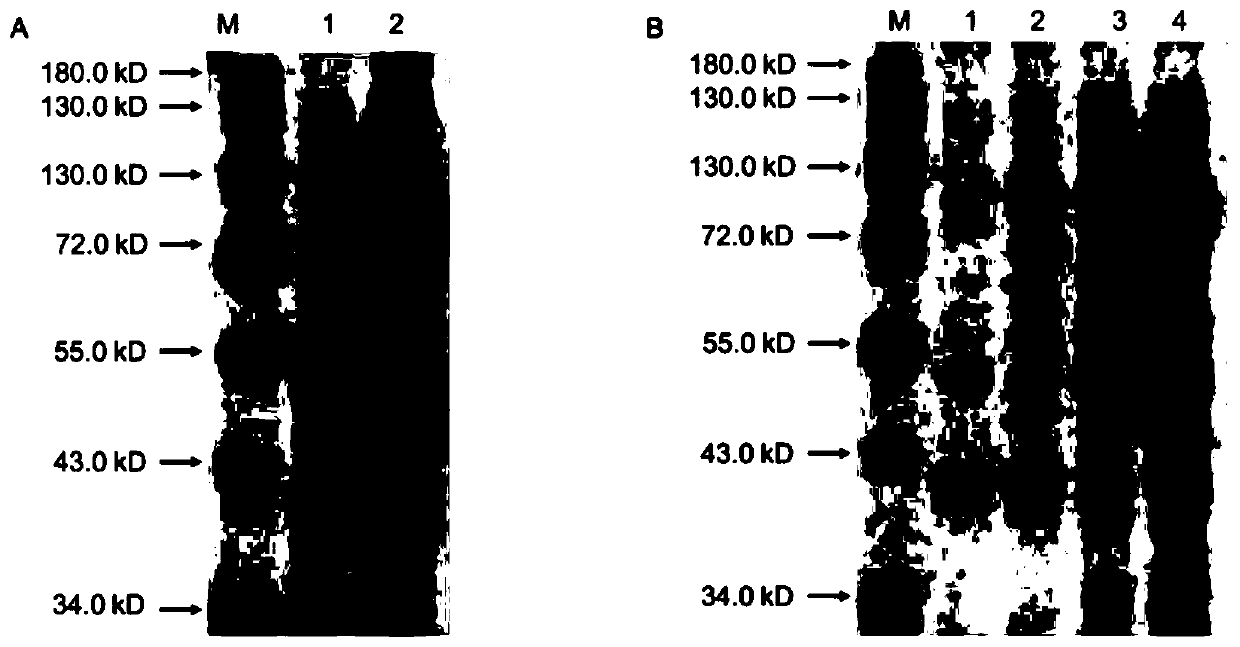
[0038] Inoculate the recombinant strain BL21 (pET32b-EST-T2) into 4ml LB liquid medium containing ampicillin (50μg / mL), culture overnight at 37°C with shaking; the next day, transfer to 100mL LB liquid medium at a ratio of 1:100 , cultured with shaking at 37°C until OD 600 When the value is 0.5, add IPTG to a final concentration of 0.2mmol / L, and induce at 30°C for 24h; centrifuge the expressed Escherichia coli at 8000rpm at 4°C, discard the supernatant, add 20mL of PBS buffer to resuspend the bacteria and resuspend The centrifugation was repeated 3 times, and finally 20 mL of PBS buffer was added to resuspend the bacteria, and they were crushed with an ultrasonic breaker in an ice-water bath (ultrasonic conditions were: ultrasonic time 2 s, intermittent time 8 s, and the total working time was set to 10 min). Complete the preparation of the crude enzyme solution. SDS-PAGE electrophoresis wa...
Embodiment 3
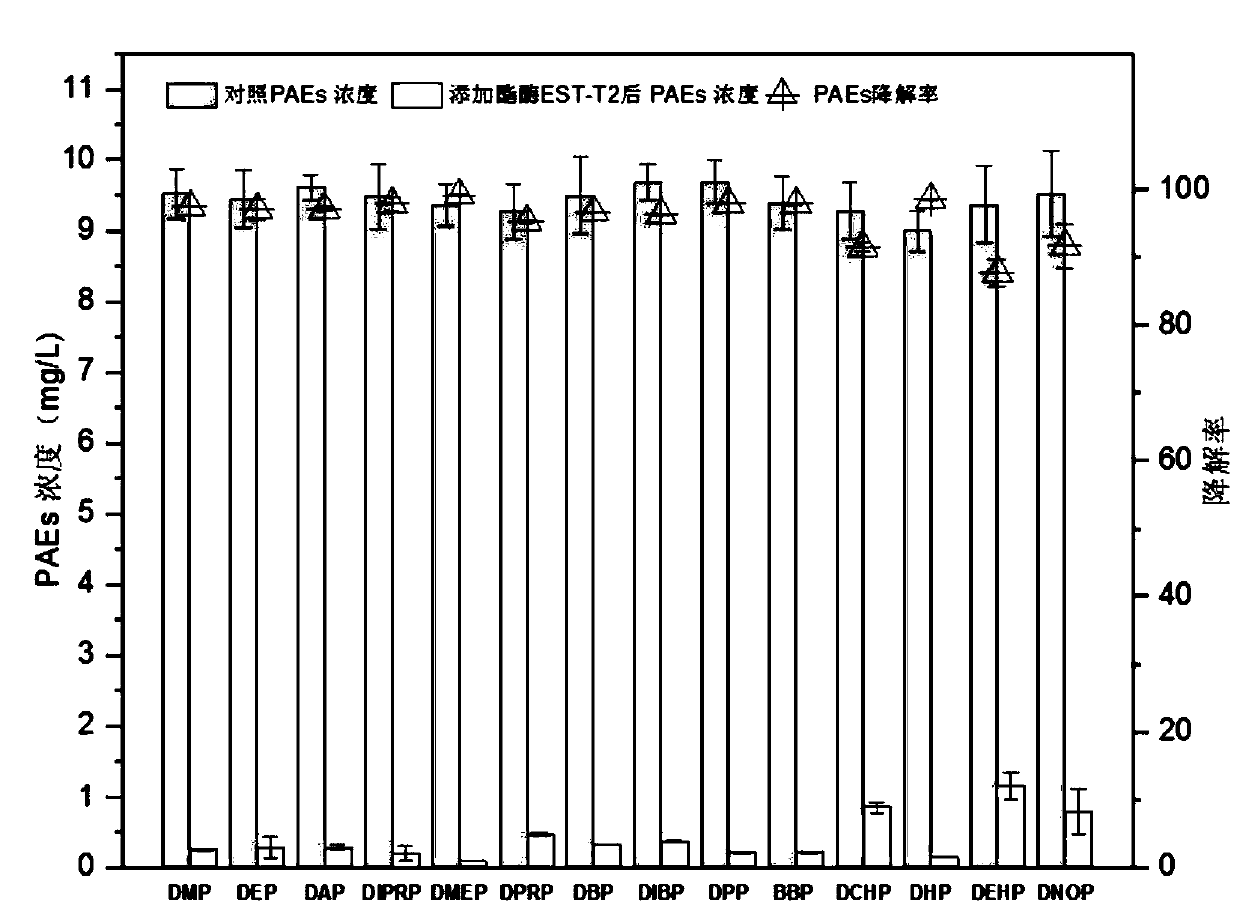
[0040] Example 3 Analysis of Esterase EST-T2 Degradation PAEs Spectrum
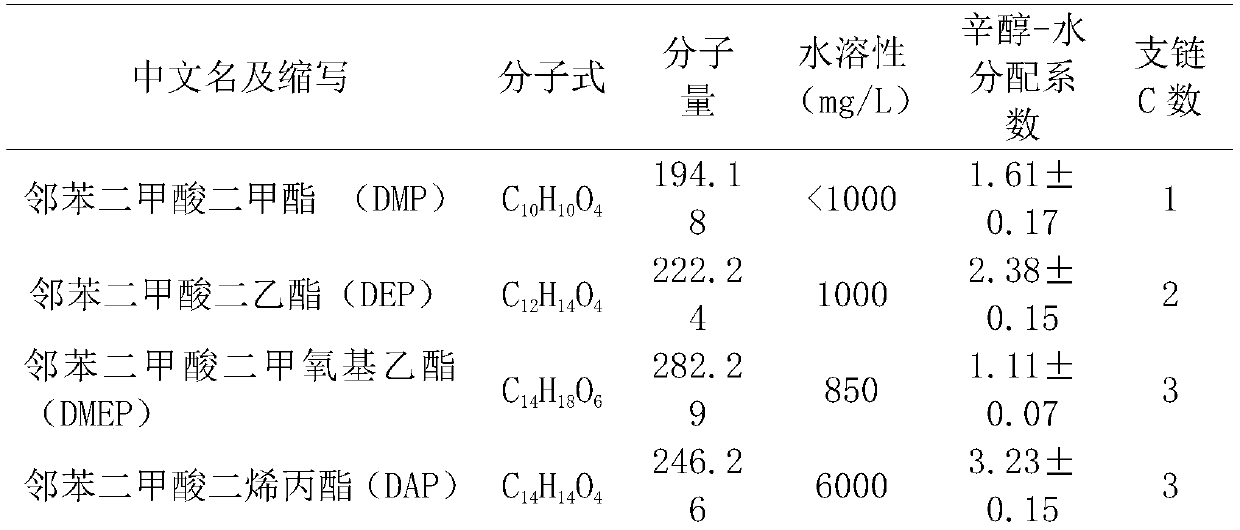
[0041] Get 1mL of the crude enzyme solution made in Example 2 and add it into 4mL of PBS buffer to form a 5mL reaction system, add a mixed solution of 14 kinds of PAEs in Table 1 dissolved in acetonitrile so that the concentration of various PAEs in the reaction system is 10mg / L , and then reacted in a constant temperature water bath at 37°C for 1 h, and verified the content of PAEs by GC-MS. The empty crude enzyme solution of pET32b was used as the blank control, and each treatment was repeated 3 times.
[0042] The extraction and analysis method of 14 kinds of PAEs: Add the same amount of acetonitrile to the reaction system to be tested, shake and extract on a shaker at 150 r / min for 40 minutes, take out and add 15gNaCl, vortex for 2min to make layers, draw 1mL supernatant into 10mL glass In a centrifuge tube, blow dry with nitrogen, dilute to 1 mL with chromatographically pure acetonitrile, dilute 10 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com