Fusion expression method of AEP cyclase in Escherichia coli, method for identifying cyclization capacity of AEP cyclase and application of APE cyclase
An Escherichia coli, fusion expression technology, applied in the field of genetic engineering, can solve the problem that the expression level cannot meet industrial production, and achieve the effect of improving stability and extending half-life
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
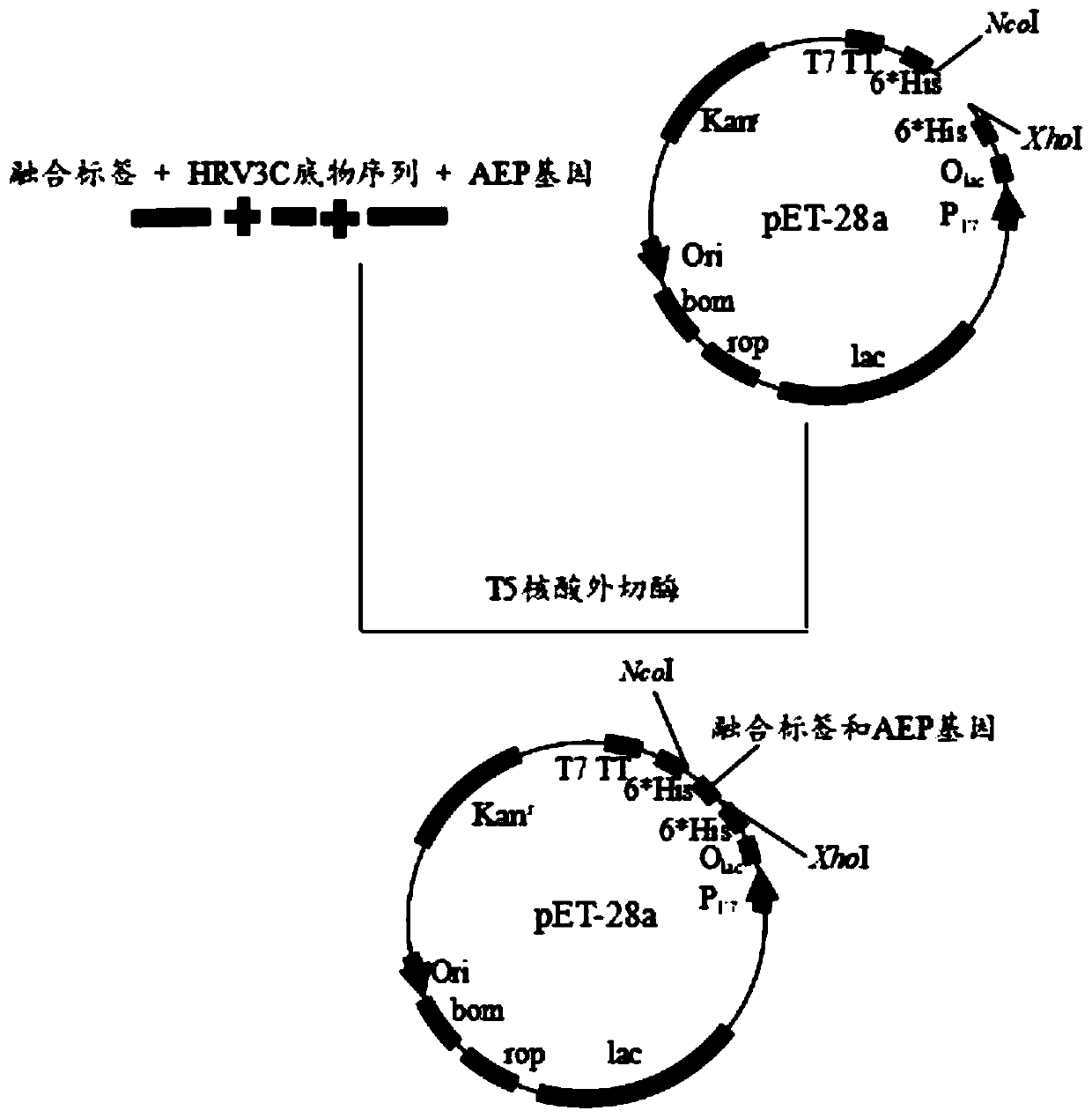
[0033] Embodiment 1 Fusion expression of AEP cyclase in Escherichia coli
[0034] In this example, five fusion tags of MBP, NusA, SUMO, GFP, and Ubiquitin were used to fuse and express AEP, and vectors containing corresponding fusion tags were constructed, which were pET28a-GFP-AEP-6*His, pET28a-MBP-AEP- 6*His, pET28a-SUMO-AEP-6*His, pET28a-NusA-AEP-6*His, and pET28a-Ubiquitin-AEP-6*His. The specific experimental plan is as follows:
[0035] S1: Link the AEP sequence containing the protease polypeptide substrate recognition sequence with the fusion tag to construct a recombinant DNA sequence.
[0036] Use the AEP fragment synthesized by the gene as a template for PCR amplification. The sequence of the AEP fragment is shown in SEQ ID NO.16. The total PCR system is 100 μL, including 10 μL of 10×pfu Buffer, 4 μL of dNTPs (10 mmol / L), forward primer and reverse 4 μL of each primer, plasmid as template: 20-30ng, finally add 4 μL of pfu DNA polymerase, and finally add water to mak...
Embodiment 2
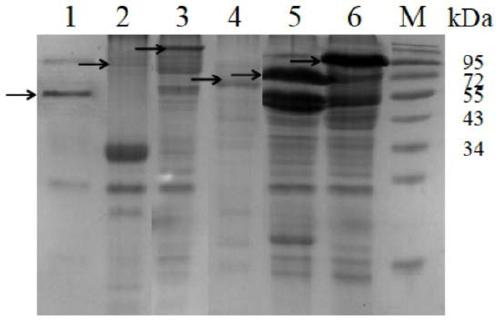
[0078] Example 2 Identification of cyclization ability of AEP cyclase
[0079] Step 1: Obtain the cyclization substrate of AEP cyclase
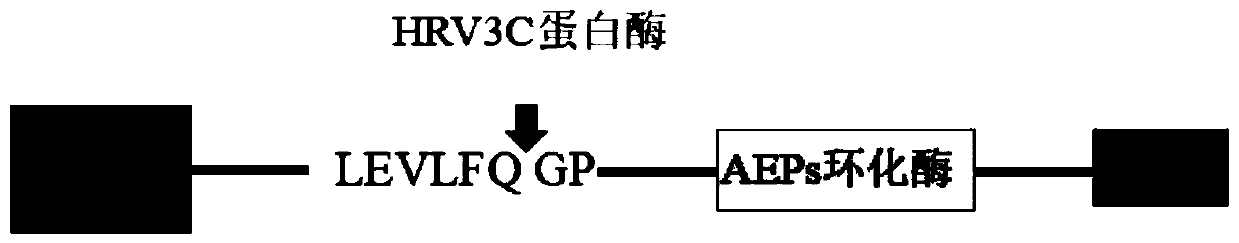
[0080] Since the SUMO protein is 11.0kDa in size, it can be well observed in SDS-PAGE and has the possibility of cyclization, so it is used as the cyclization substrate of AEP cyclase. Add GGGGSGGGGS to the N-terminal of the SUMO protease substrate, and add a longer Linker1 (GSGS), TEV protease substrate sequence (ENLYFQ / S), and a longer Linker2 (DVGGGGSEGGGSGGPGSGGEGSAGGGSAGGGS) to the C-terminal of the SUMO protease substrate in sequence and 6*His(HHHHHH), and finally add the recognition sequence of AEP cyclase at both ends to form a tandem substrate (GLP-SUMO protease substrate-Linker1-TEV protease substrate-Linker2-STRN / GLP-6*His ), the recognition sequence of AEP cyclase is shown in SEQ ID NO:13. Tandem substrates were constructed by overlap extension PCR, and the primer sequences were as follows:
[0081] SUMO-TEV-F: aactttaagaaggaga...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com