Genome editing method based on zymomonas mobilis endogenous CRISPR-Cas system and application thereof
A Zymomonas, genome editing technology, applied in the field of genetic engineering, can solve the problems of host large cells, toxicity, nuclease inactivity, etc., to avoid cytotoxicity, strong positive selection pressure, and avoid limited available selection markers.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Construction of Zymomonas mobilis Endogenous CRISPR-Cas Genome Editing System
[0040] (1) CRISPR-Cas phylogenetic analysis of the genome encoding of Zymomonas mobilis
[0041] The premise of the present invention is that the research strain needs to encode the CRISPR-Cas system itself and have DNA splicing activity, which requires the host genome to encode the CRISPR cluster and the complete Cas protein system.
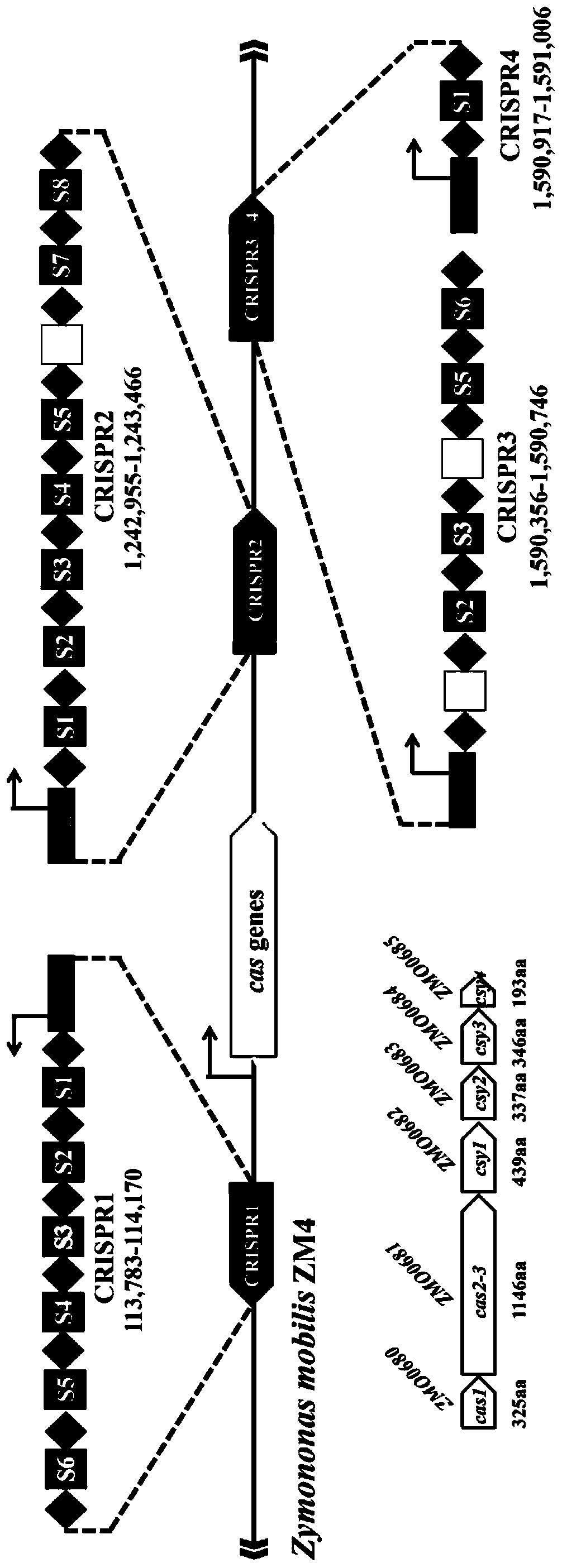
[0042]Taking Zymomonas mobilis Z.mobilis 4 as the model strain, the sequencing data of Z.mobilis 4 were analyzed. The results showed that the genome of the strain encoded four CRISPR structural sequences. According to their sequence in the genome, we sequenced them named CRISPR1-CRISPR4, see figure 1 . CRISPR1 occupies the 113,783-114,170 region of the genome and contains 7 repeat sequences; CRISPR2 occupies the 1,244,355-1,245,866 region and contains 9 repeat sequences; CRISPR3 occupies the 1,598,754-1,599,144 region and contains 7 repeat sequence...
Embodiment 2
[0074] Example 2 Application of Genome Editing Method Based on Zymomonas mobilis Endogenous CRISPR-Cas System in Gene Knockout
[0075] In this embodiment, the selected target gene is the ZMO0038 gene encoded by the Zymomonas mobilis genome, and the specific steps are as follows:
[0076] (1) GuideRNA sequence selection: The 32 bp sequence immediately downstream of any 5'-CCC-3' was intercepted from the target gene sequence ZMO0038 as a guideRNA, and the sequence could be located on any strand of the genome.
[0077] ZMO0038-guideRNA primer sequence:
[0078] 0038-gRNA1-F: GAAAATGCCGCTTTCGATCAGGCCGCCGCCAAGATT, see SEQ ID NO:9;
[0079] 0038-gRNA1-R: GAACAATCTTGGCGGCGGCCTGATCGAAAGCGGCAT, see SEQ ID NO: 10;
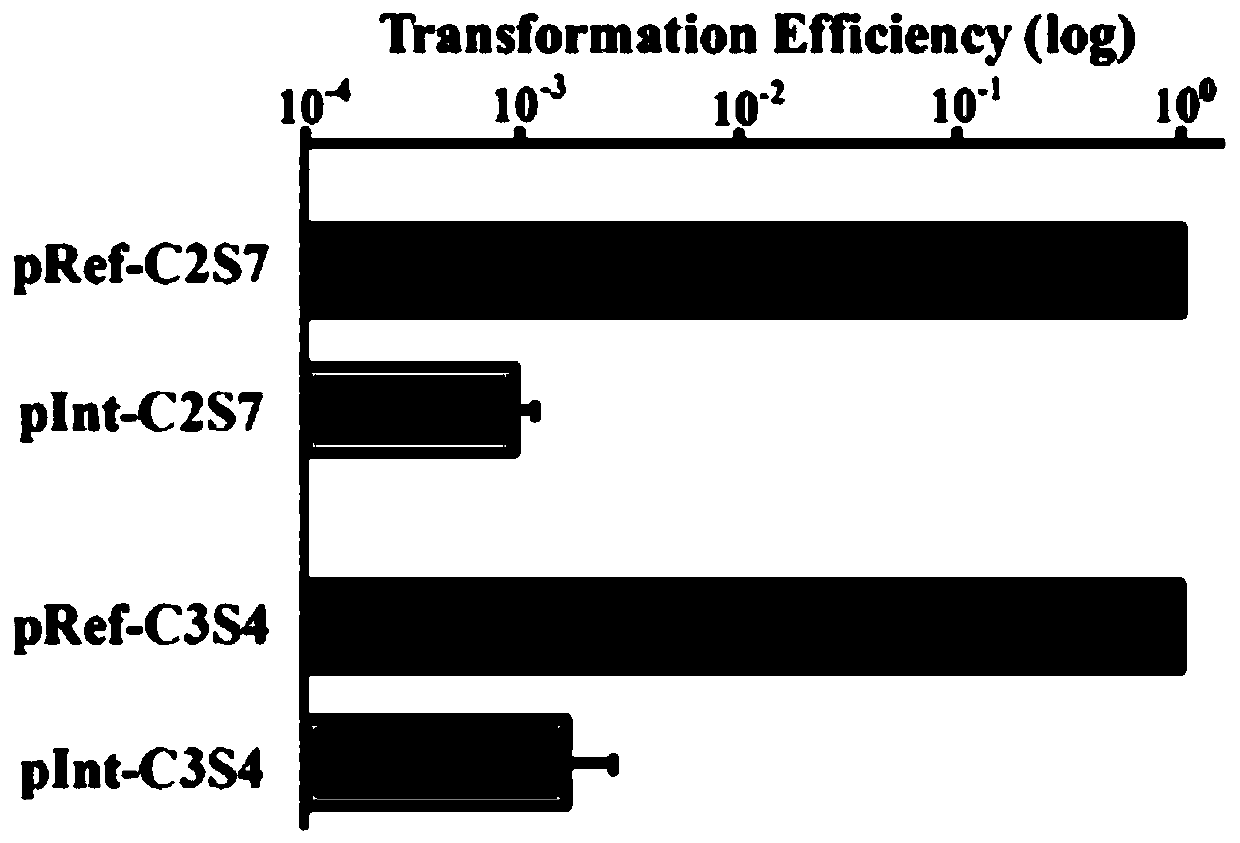
[0080] (2) guideRNA primer sequence is constructed on the vector, see Figure 5 : The above-mentioned plasmid containing the artificial CRISPR expression unit was linearized with BsaI (Table 5), annealed with the guideRNA primer (same as above, Table 2) and ligated with ...
Embodiment 3
[0103] Example 3 Application of Genome Editing Method Based on Zymomonas mobilis Endogenous CRISPR-Cas System in Site-directed Insertion of DNA Sequence
[0104] In this embodiment, a His-Tag tag is inserted at a fixed point after the start code ATG of the target gene ZMO0038, see Figure 8 , so that the encoded protein can be purified through a nickel column. This method can be extended to any protein to be purified. Specific steps are as follows:
[0105] (1) ZMO0038(His-Tag)-guideRNA sequence selection: intercept the 32 bp sequence immediately downstream of the 5'-TCC-3' near the start codon ATG of the target gene sequence ZMO0038 as a guideRNA, and this sequence can be located on any strand of the genome.
[0106] ZMO0038(His-Tag)-guideRNA primer sequence:
[0107] 0038His-gRNA-F: GAAAATCTTGACTCCCTCCATGCACTTAAAAAATCT, see SEQ ID NO: 18;
[0108] 0038 His-gRNA-R: GAACAGATTTTTTAAGTGCATGGAGGGAGTCAAGAT, see SEQ ID NO:19.
[0109] (2) GuideRNA is constructed on the vector:...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com