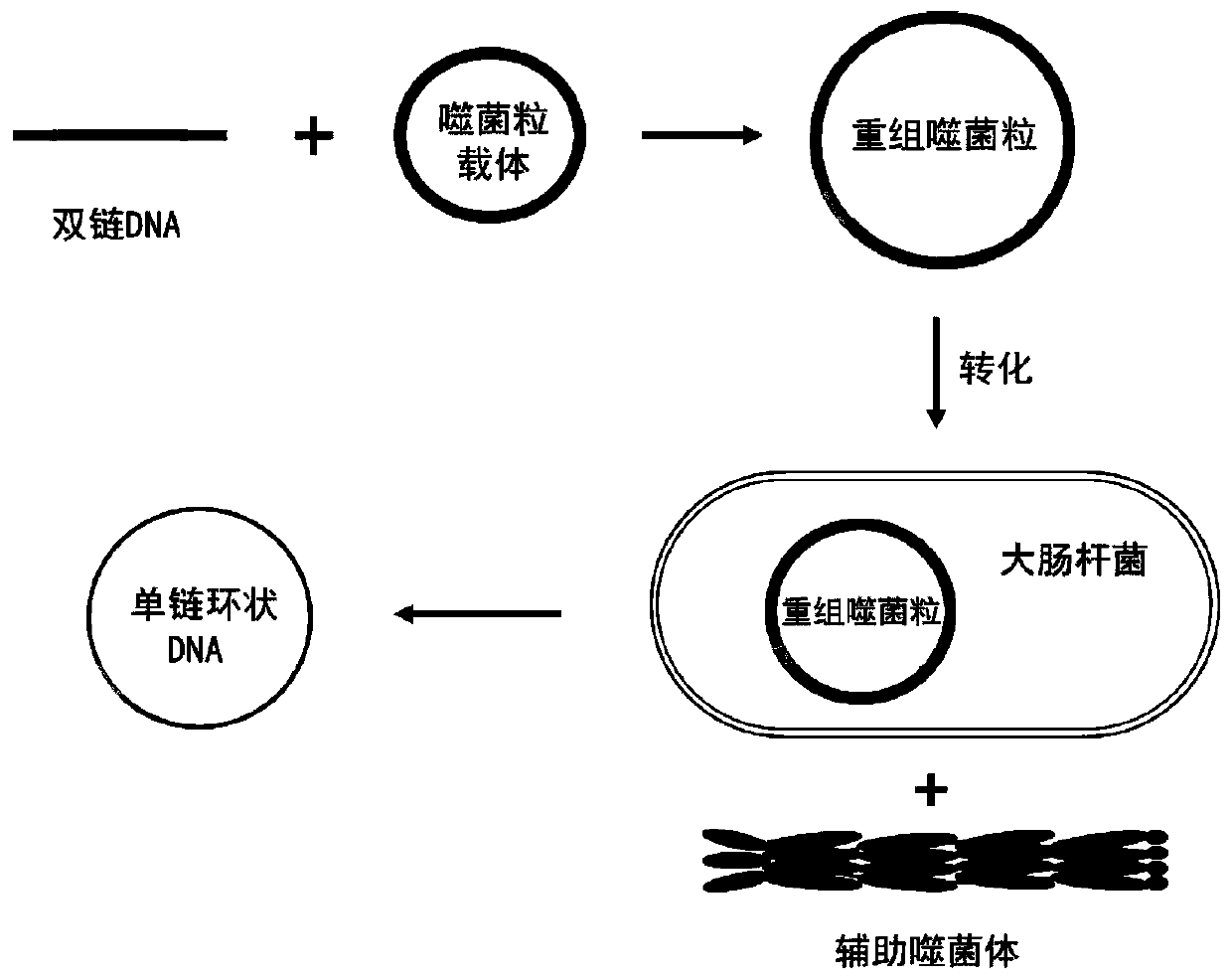
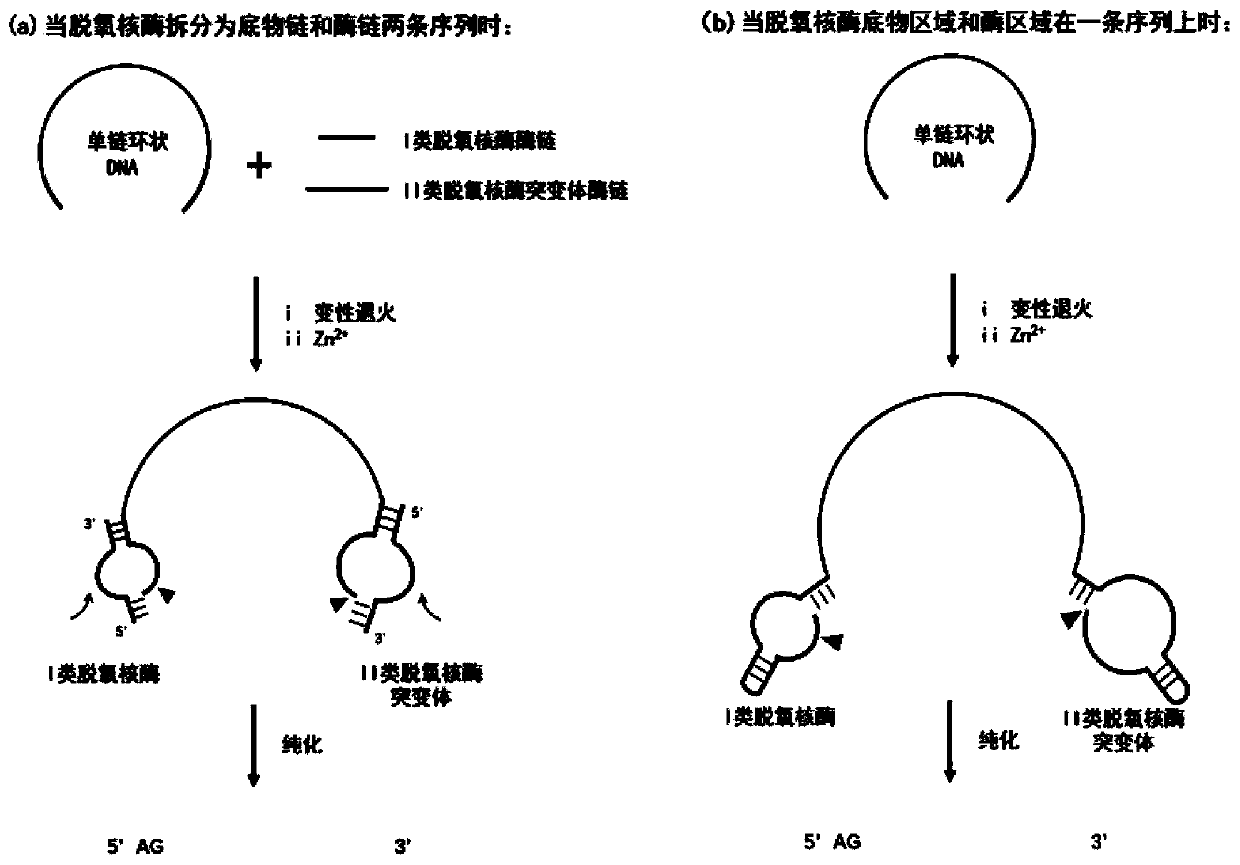
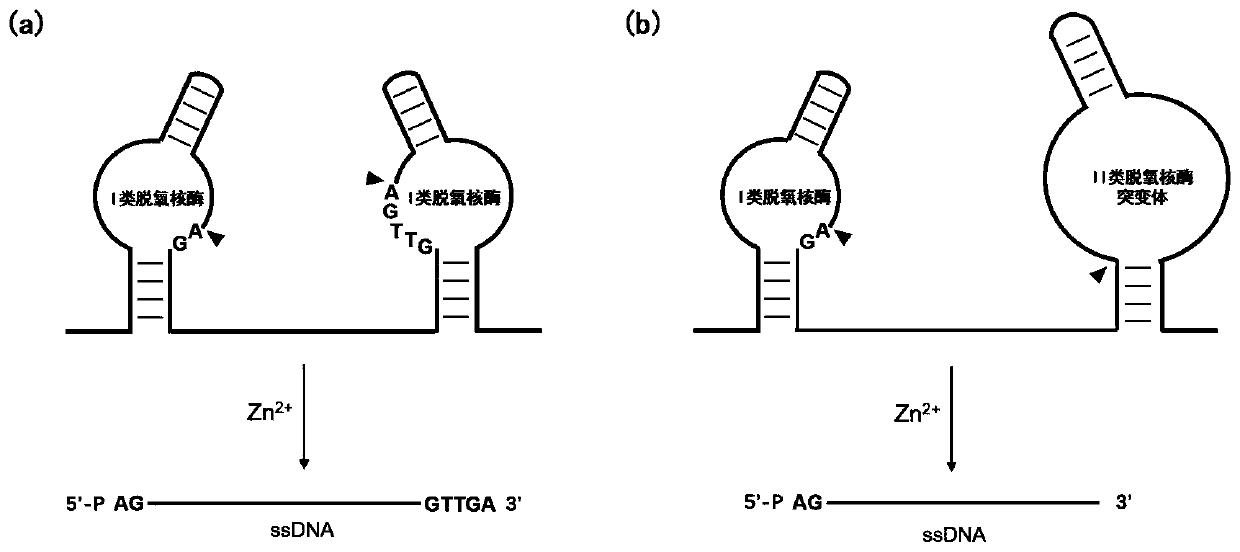
Preparation method of long single-chain DNA
A single-strand, sequence technology, applied in the field of preparation of long single-strand DNA, can solve the problem that the single-strand DNA sequence cannot be customized, and achieve the effect of efficient cutting, high-efficiency preparation, and high purity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Deoxyribozyme combined with helper phage to prepare single-stranded DNA
[0035] Using mEGFP as a template, PCR primers were designed to amplify the target fragment.
[0036] Such as figure 2 As shown in (a), when the deoxyribozyme is split into a substrate chain and an enzyme chain, and when cutting occurs in the form of two sequences, the class I deoxyribozyme substrate sequence is added to the forward primer, and the reverse The substrate sequence of class II deoxyribozyme mutants was added to the primers, and on this basis, BamH I and Hind III restriction sites were added to the 5' ends of the forward and reverse primers, respectively. Wherein, according to the last base at the 3' end of the single-stranded sequence to be prepared, the corresponding class II deoxyribozyme mutant is selected. If the last base at the 3' end is G, select II-R1a; if the last base at the 3' end is A, select II-R1b; if the last base at the 3' end is T, select II-R1c; such as ...
Embodiment 2
[0055] Taking 60nt and 160nt sequences as examples, the purity of single-stranded DNA prepared by the present invention was compared with that of chemically synthesized single-stranded DNA.
[0056] The method described in Example 1 was used to prepare 60 nt single-stranded DNA. The same sequence chemically synthesized was ordered from the company, and the purification method was polyacrylamide gel purification. A part of the sample was purified by polyacrylamide gel again in the laboratory. By performing 12% polyacrylamide gel (Acr / Bis 19:1) electrophoresis, the purity of the single-chain sample prepared by this method and the single-chain sample after purification by chemical synthesis was compared. The result is as Figure 5 Shown in (a), swimming lane A is the 60nt single strand obtained by chemical synthesis and purification once, swimming lane B is the 60nt single strand obtained by chemical synthesis and purification twice, and swimming lane C is the 60nt single stran...
Embodiment 3
[0059] The knock in experiment of mEGFP targeting microtubule TUBA1B gene, the experimental principle is as follows Figure 6 shown. The single-stranded DNA with a length of 1570 nt prepared in Example 1 of the present invention is used as a single-stranded DNA repair template, and its application in the knock in experiment. Specific steps are as follows,
[0060] 1. Cell culture. Hek293T cells were purchased from the Cell Bank of Shanghai Type Culture Collection Committee, Chinese Academy of Sciences. The culture condition is DMEM medium containing 10% FBS (inactivated), which contains 50 units / mL of penicillin, 50 μg / mL of streptomycin, and 4 mM of glutamine. Place at 37°C, 5% CO 2 Concentration in a constant temperature incubator.
[0061] 2. Construction of CRISPR / Cas9 plasmid vector targeting microtubule TUBA1B gene.
[0062](1) Synthesis of sgRNA double-stranded fragments. The sgRNA sequence targeting the human TUBA1B gene is TGGAGATGCACTCACGCTGC (SEQ ID NO: 16) (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com