SNP markers for distinguishing resistance to Vibrio harveyi infection of Litopenaeus vannamei and its detection method and application
A vannamei and molecular marker technology, applied in the SNP marker for distinguishing the anti-Vibrio harvetii infection ability of Litopenaeus vannamei and its detection and application fields, can solve the problems of cumbersome operation, high cost and the like, and achieve the effect of low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
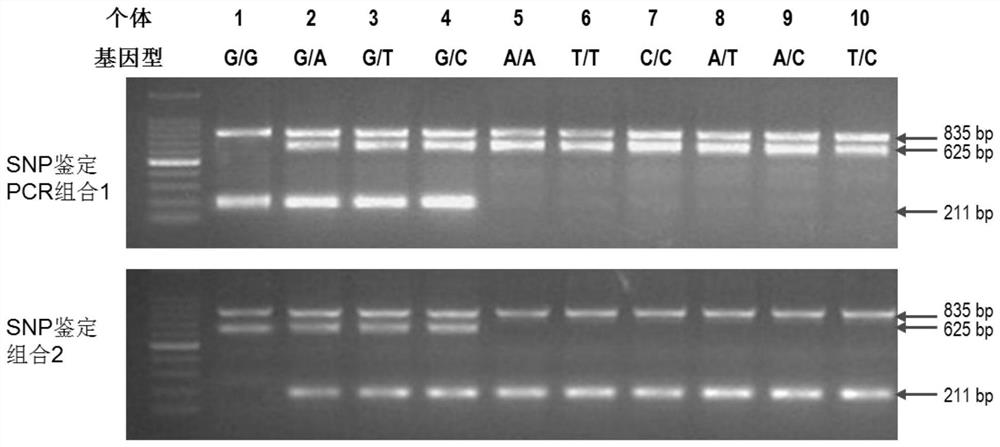
[0040] A prawn farm in Zhanjiang had 1,000 tails of Litopenaeus vannamei with a size of 8-10cm. Swimming feet were taken to extract the genomic DNA of each Litopenaeus vannamei (taking the tissue does not affect the survival of the Litopenaeus vannamei), and carried out the Lyz- i2 gene four-primer amplification hindered mutation system region PCR detection (determining the genotyping of the SNP site), the specific method is as follows:
[0041] 1. Amplification primers:
[0042] LyzSNPF1: 5'-GAGACCGTCCGCCGCTACAT-3'
[0043] LyzSNPR1: 5'-ACCACAAACGACACCCTACCATT-3'
[0044] LyzSNPF2: 5'-TCAACTCCTCACGCGAGTAG-3';
[0045] LyzSNPR2: 5'-GCCGTTGCCGTCGCAATCD-3';
[0046] LyzSNPF3: 5'-TCAACTCCTCACGCGAGTAH-3';
[0047] LyzSNPR3: 5'-GCCGTTGCCGTCGCAATCC-3';
[0048] Among them, D is A, T, G degeneracy, H is A, T, C degeneracy.
[0049] 2. Amplification system:
[0050] (1) Amplification system combination 1:
[0051]
[0052] (2) Amplification system combination 2:
[0053] ...
Embodiment 3
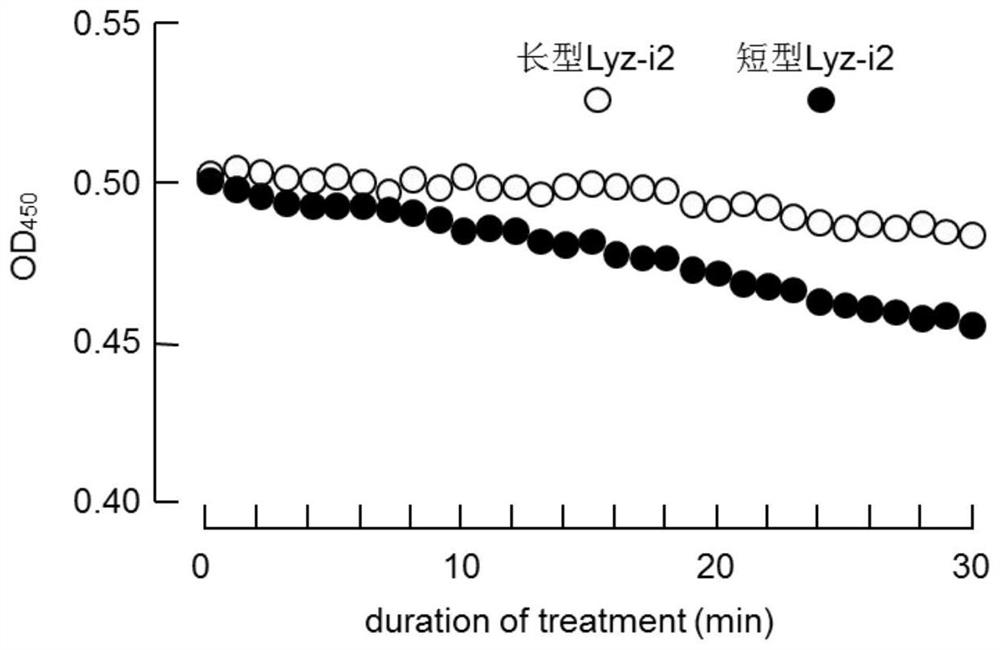
[0062]Vibrio harveyi was isolated from the hepatopancreas of luminescent vibrio prawn Litopenaeus vannamei by the South China Sea Institute of Oceanology, Chinese Academy of Sciences. The strains were stored in 25% glycerol at -80°C, streaked in LB solid medium, inoculated in LB liquid medium and cultured with shaking at 30°C for 24 hours. The bacterial solution was centrifuged at 3000 g for 10 minutes at high speed, and the precipitated bacterial cells were washed 3 times with PBS buffer and then suspended in an equal volume. The concentration of the bacterial solution was determined by a spectrophotometer at 560nm (OD560).
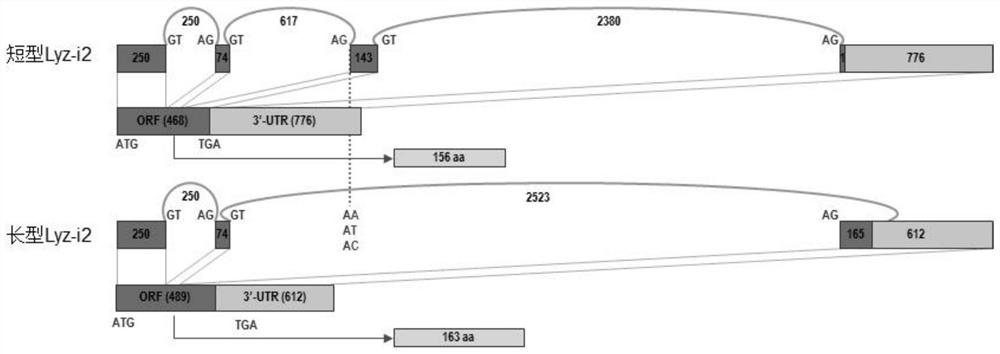
[0063] Then, PBS (50 mM, pH 6.2) was used to resuspend to prepare a Vibrio harveyi suspension, which was measured by a spectrophotometer at 450 nm (OD450), so that its OD450 was 0.6. The long and short Lyz-i2 recombinant proteins (the short Lyz-i2 protein sequence is shown in SEQ ID NO.3, and the long Lyz-i2 protein sequence is shown in SEQ ID NO.5) wer...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com