High-throughput identification method of internal ribosome entry site (IRES) elements in various source cell samples
A technology of binding sites and identification methods, applied in nucleic acid vectors, botanical equipment and methods, biochemical equipment and methods, etc., can solve the problems of unreal results, increased capital investment, low throughput, etc., and achieve a clear purpose , improve the accuracy, the effect of the process is simple
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
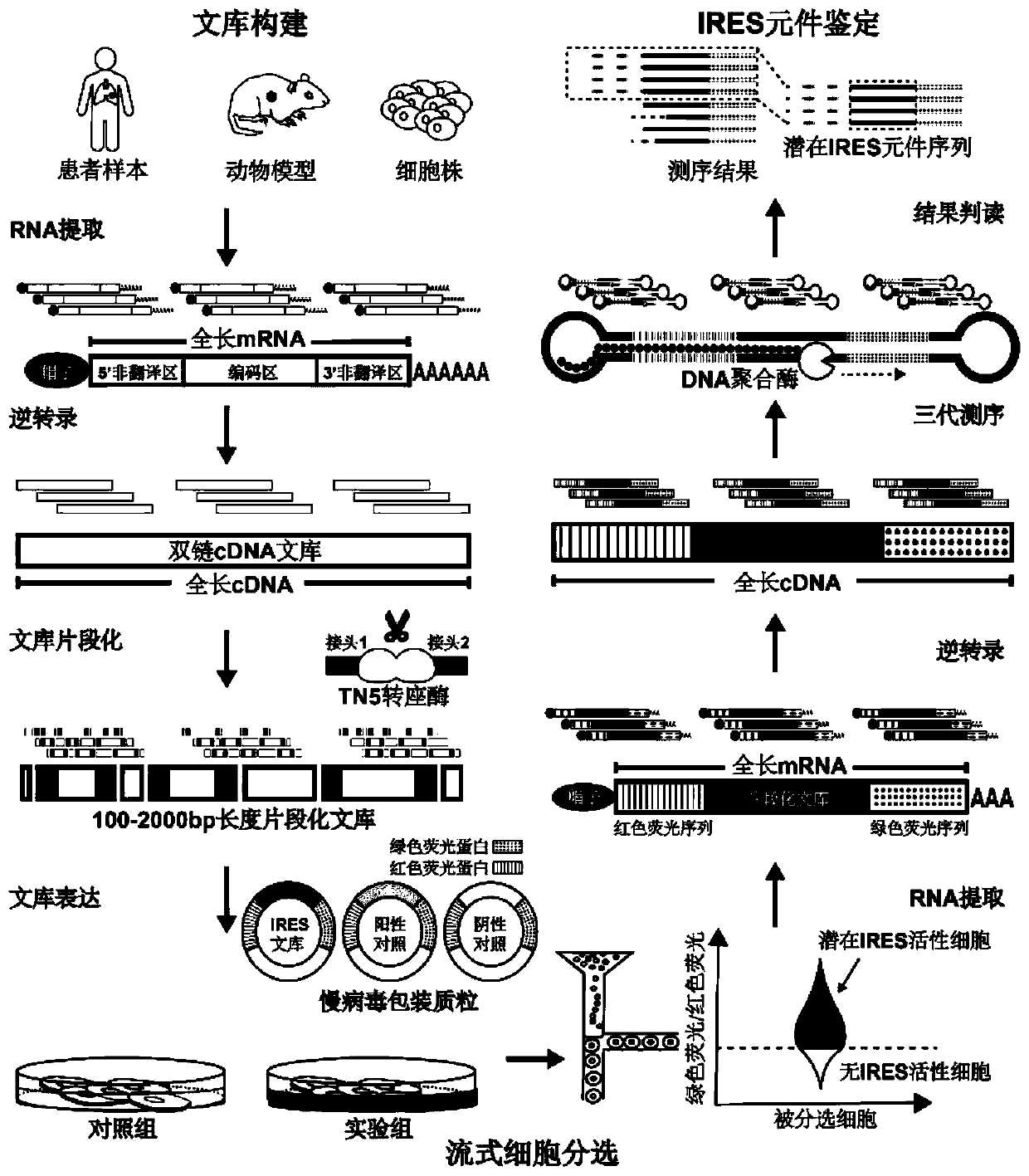
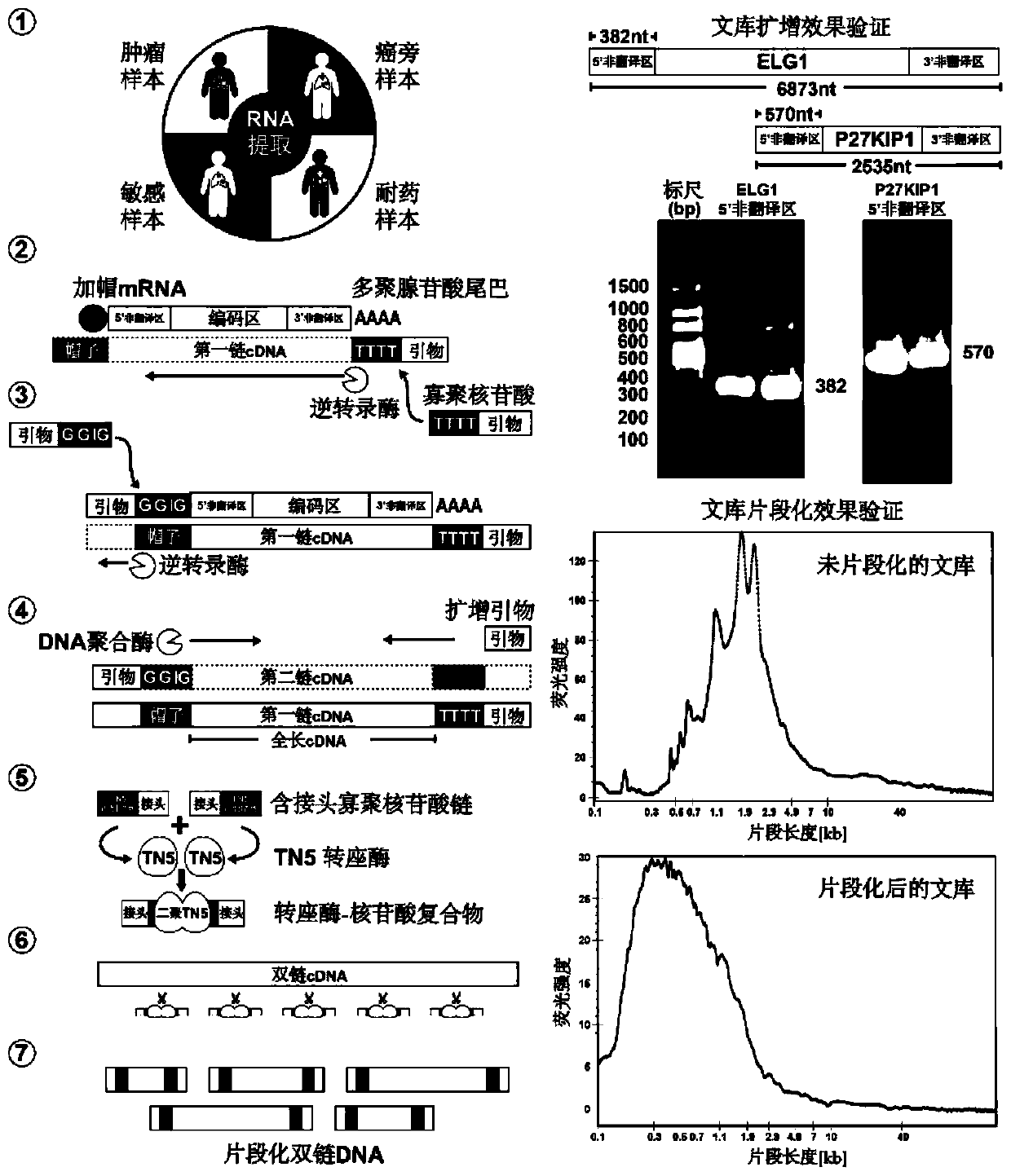
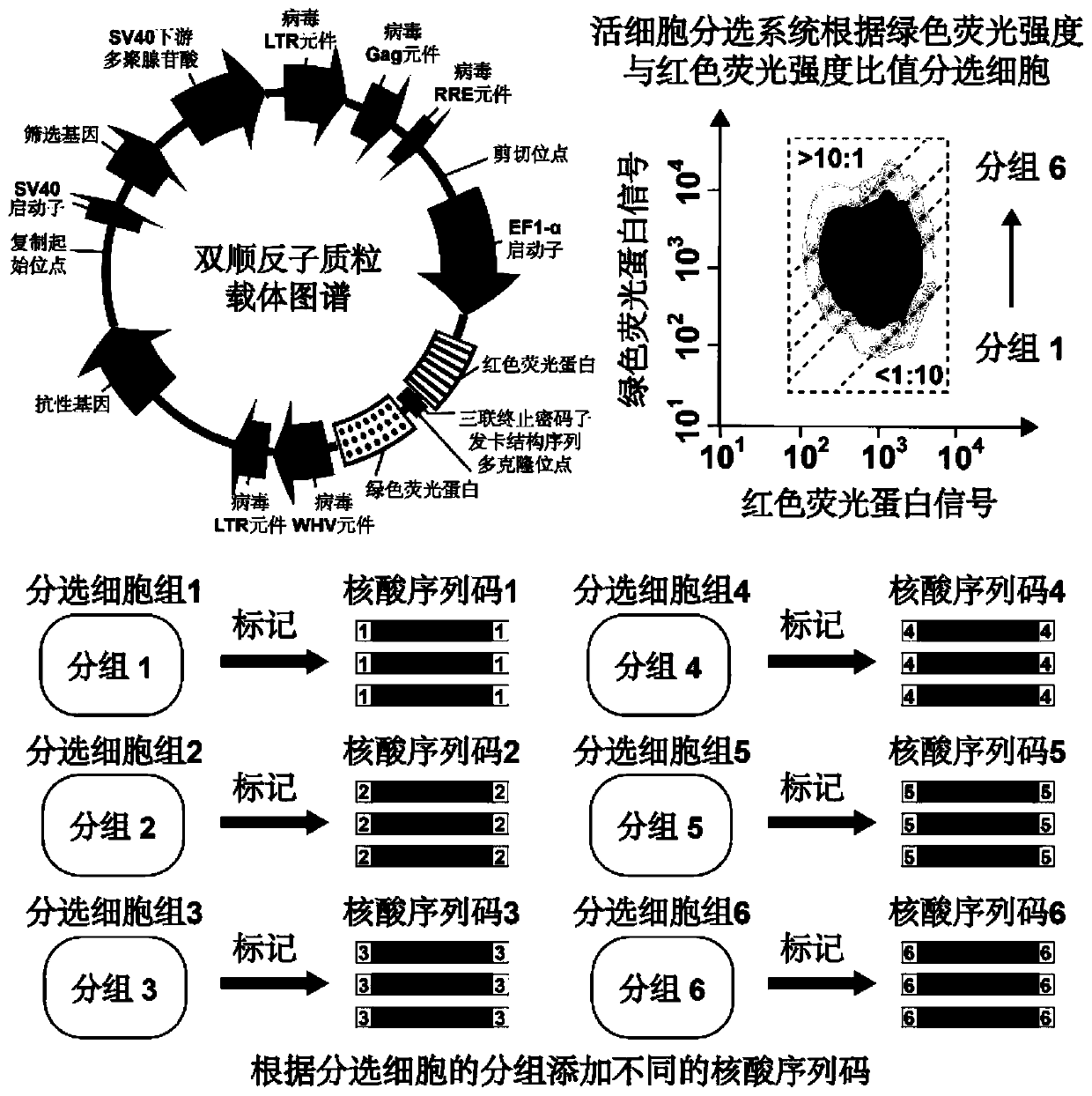
[0037] The high-throughput identification method for internal ribosome binding site elements in cell samples from multiple sources comprises the following steps:
[0038]1. Use the RNA extraction reagent Trizol to extract the total RNA from cells. Use the Nanodrop spectrophotometer to detect the concentration of the extracted total RNA, and use two oligomer cores designed to bind to the 5'-end cap structure and the 3'-end polyadenylic acid tail sequence on the mRNA chain respectively The nucleotide chain undergoes reverse transcription. The sequence of the oligonucleotide chain is: an oligonucleotide (5'-3') capable of binding to the 5'-end cap structure: AAGCAGTGGTATCAACGCAGAGTACATGGG (SEQ ID NO.1; wherein, the penultimate and penultimate Three Gs are ribonucleotide bases G; the last G is a locked nucleotide base G); the rest are composed of deoxyribonucleotide bases marked in the sequence), which can bind to the 3'-terminal polyadenylation Oligonucleotide (5'-3') of nucleo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com