Optimized culture method and application of butyribacterium methylotrophium
A technology of methylbutyric acid bacteria and methyl butyric acid bacteria, applied in the field of microbiology, can solve problems such as difficulty in accepting foreign genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
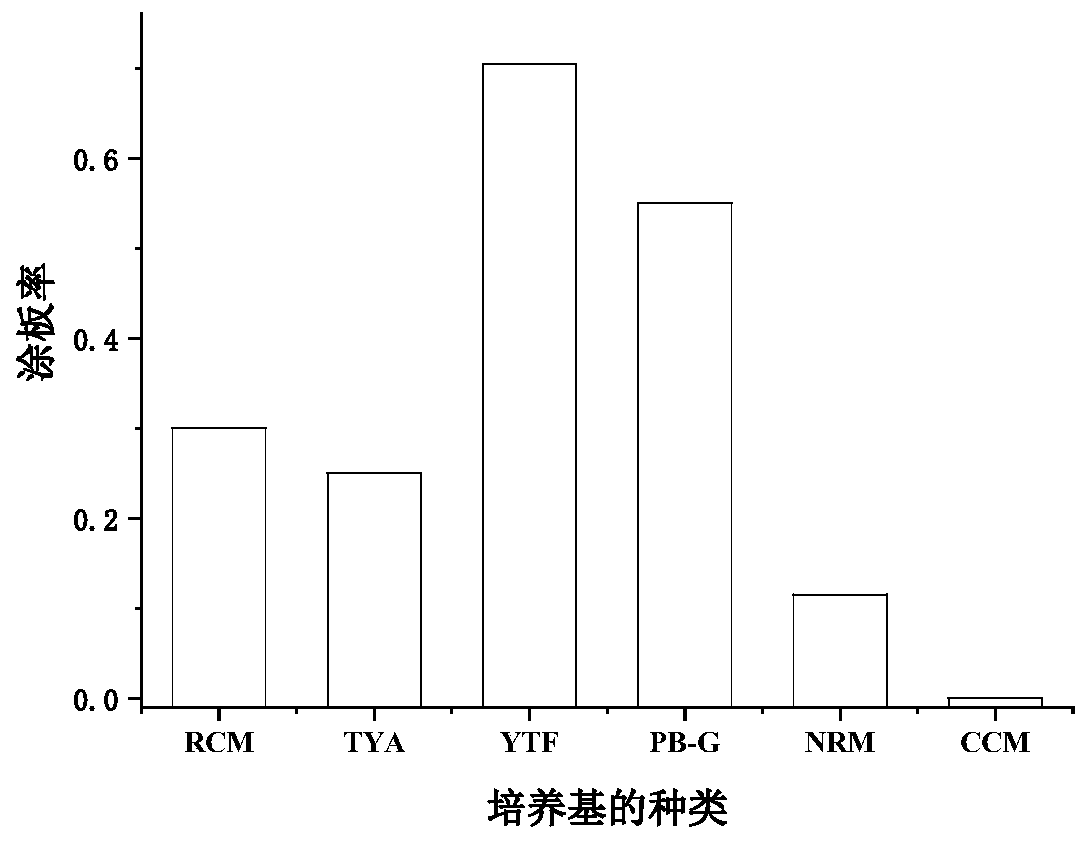
[0030] Example 1 Screening different types of medium
[0031] Six media suitable for the growth of Clostridia were selected, including
[0032] RCM (yeast extract 3g / L, beef extract 10 g / L, peptone 10 g / L, soluble starch 1 g / L, glucose 5 g / L, sodium chloride 3 g / L, sodium acetate 3 g / L),
[0033] TYA (glucose 40 g / L, beef extract 2 g / L, yeast extract 2 g / L, tryptone 6 g / L, ammonium acetate 3 g / L, potassium dihydrogen phosphate 0.5 g / L, magnesium sulfate heptahydrate 0.2 g / L, ferrous sulfate heptahydrate 0.01 g / L),
[0034] YTF (peptone 16 g / L, yeast powder 12 g / L, sodium chloride 4 g / L, glucose 5 g / L), PB-G (potassium dihydrogen phosphate 4 g / L, dipotassium hydrogen phosphate 6 g / L , ammonium chloride 1 g / L, magnesium chloride hexahydrate 0.1 g / L, calcium chloride dihydrate 0.1 g / L, yeast powder 3 g / L),
[0035] NRM (glucose 40 g / L, beef extract 2 g / L, yeast extract 2 g / L, tryptone 6 g / L, ammonium acetate 3 g / L, potassium dihydrogen phosphate 0.5 g / L, magnesium sulfate hept...
Embodiment 2
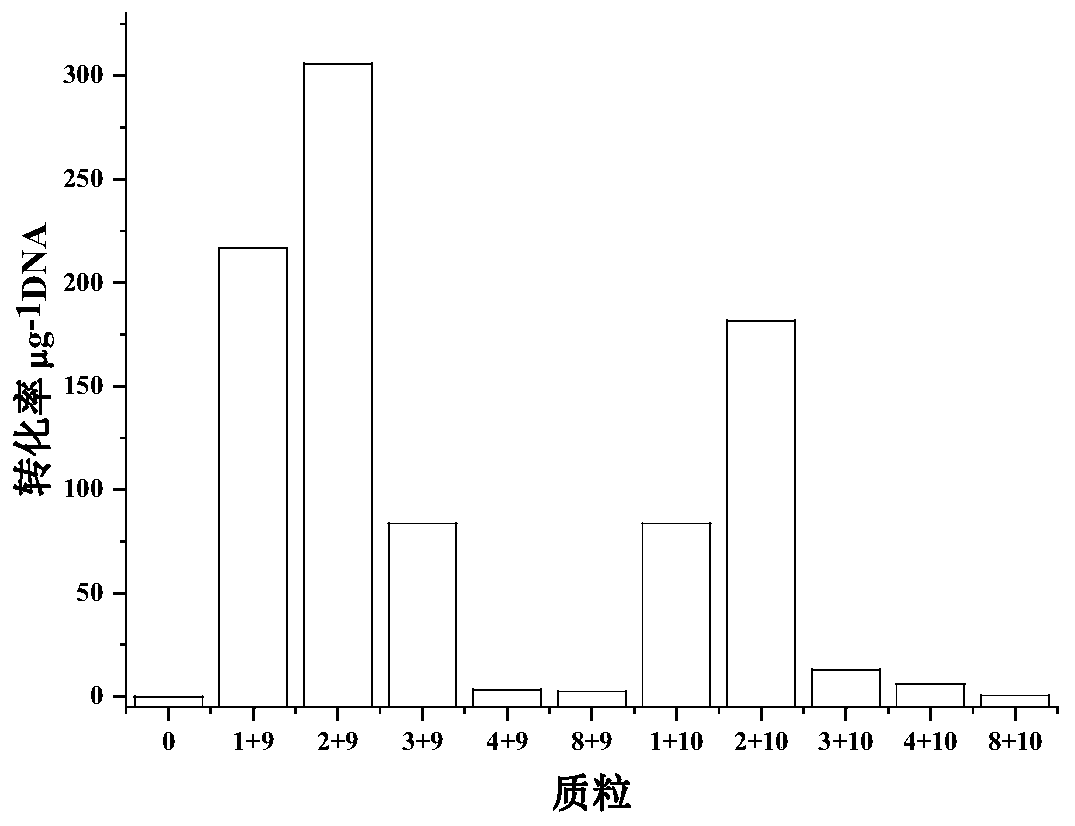
[0040] Example 2 Screening for different types of replicons
[0041] Select 5 kinds of replicons, including pIM13, pCB102, pBP1, pCD6, pIP404, corresponding to plasmid numbers 1, 2, 3, 4, 8, and select the plasmid TOP10 competent or phage containing a type 1 methylase gene to infect Bacillus subtilis The plasmid TOP10 competent for the methylase gene obtained from the bacillus, numbers 9 and 10. (The method of TOP10 competence containing the corresponding methylated gene is as follows: Transform the plasmid containing the methylase gene into commercial TOP10 competence, and spread it on solid culture. Pick a single colony from the plate Insert into a 50mL centrifuge tube containing 10mL LB culture medium, cultivate to OD at 37°C 600 After = 0.4, place the bacterial solution on ice, apply ice for 10 minutes, and centrifuge at 4000r / min for 10 minutes. Discard the supernatant, resuspend each pellet with 10ml of ice-cold 0.1mM CaCl2, place on ice for 15min, and centrifuge at 40...
Embodiment 3
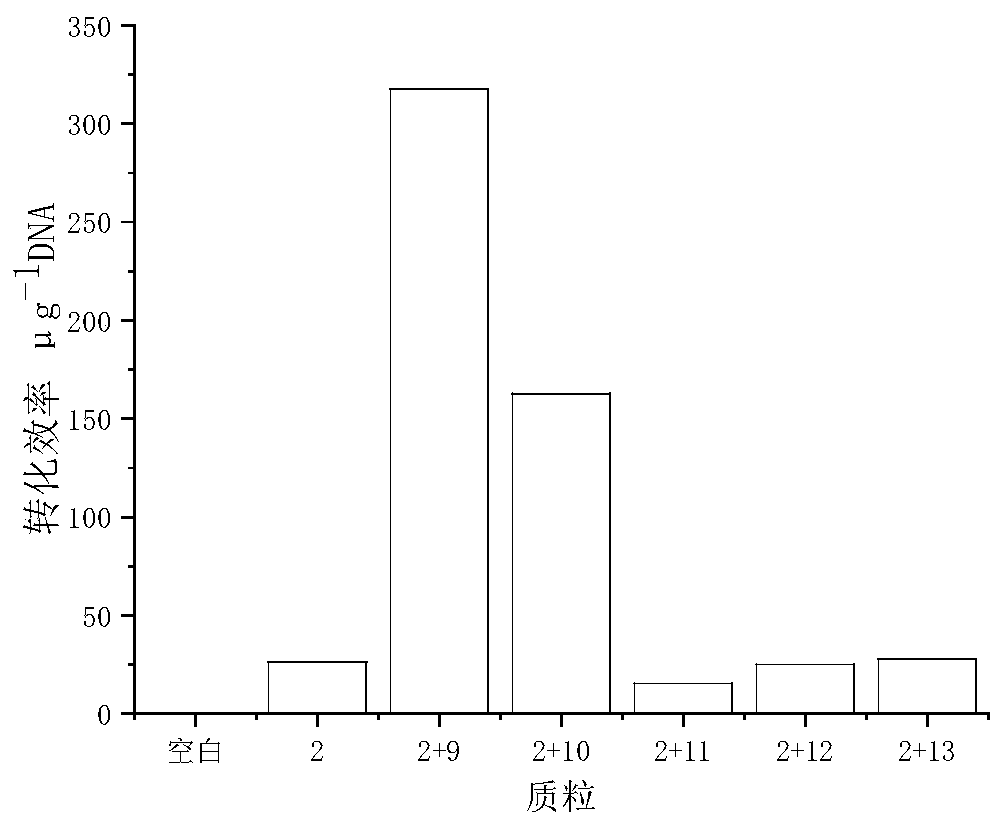
[0046] Example 3 Screening of different methylases
[0047] In order to investigate the effect of different methylases on plasmid transformation, the three types of methylases including type 1 methylase, the methylase obtained by phage infection of Bacillus subtilis, and the bacteria itself were screened. The corresponding plasmids for the enzymes are pMCljS, pAN2, pACYC184-M1, pACYC184-M2, and pACYC184-M3, respectively, and TOP10 competent cells containing the above plasmids were prepared, numbered 9-13. The most stable replicons screened were selected for electroporation experiments.
[0048] Add the plasmid containing the pCB102 replicon into TOP10 competent cells containing the methylated plasmids No. Add 1mL LB culture solution to the ultra-clean workbench, place it in a shaker at 37°C for 1 hour, and then take it out and spread it on a solid plate.
[0049] Pick a single colony from the solid plate and transfer it into a 50mL centrifuge tube containing 10mL LB culture ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com