DNA assembly method and application thereof
A purpose, recombinant bacteria technology, applied in the field of DNA assembly, can solve problems such as increasing the difficulty of DNA sequence, affecting DNA sequence, and restricting applications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
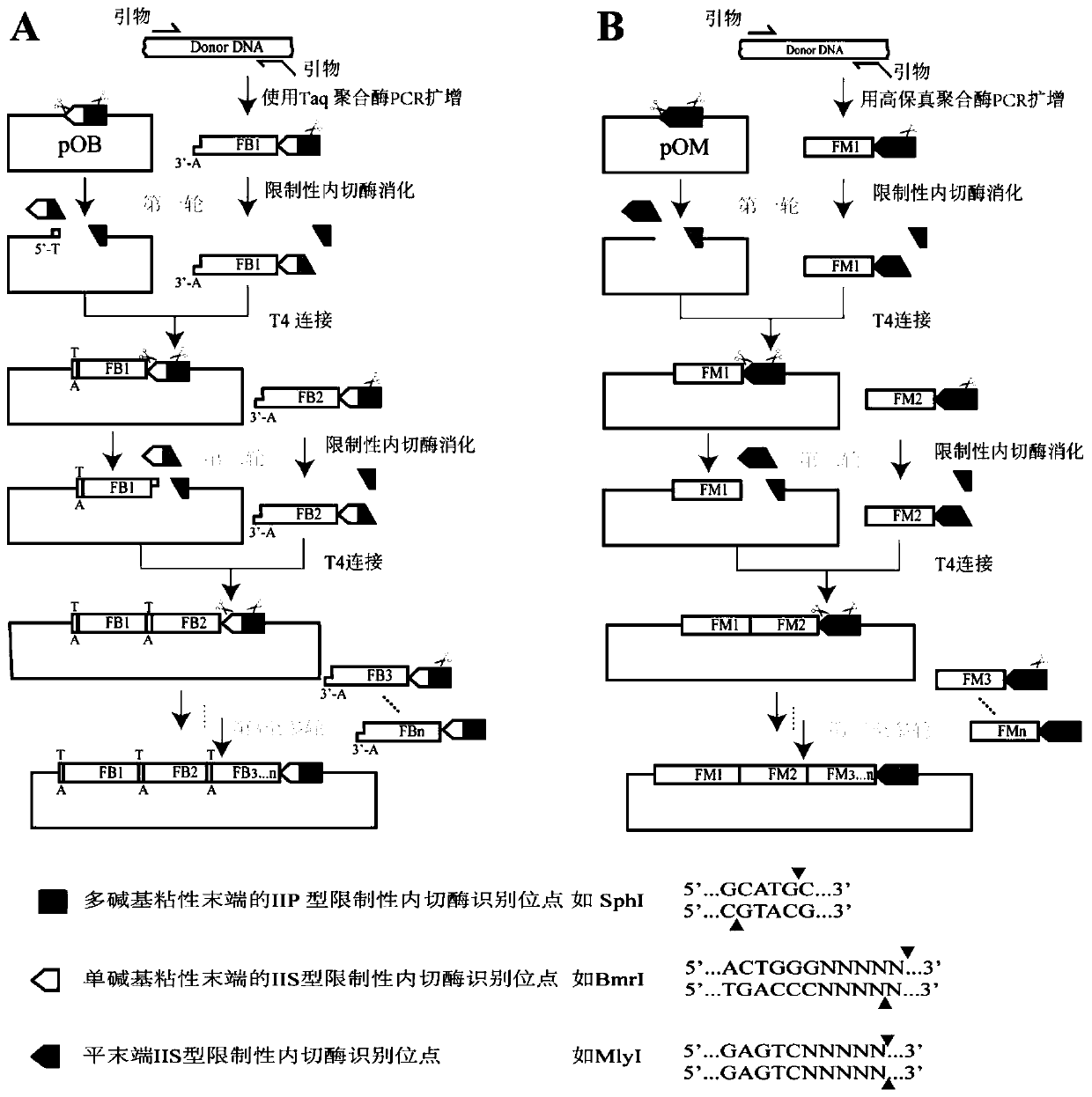
[0055] (1) Construction of PS-Brick original vector pOB plasmid and pOM plasmid
[0056] Plasmid pUC19 (SEQ ID NO.1) is used as the basic vector for verifying the PS-Brick assembly method, and a BmrI site and three MlyI sites in the vector are detected by overlap extension PCR (overlap extension polymerase chain reaction, overlap PCR) method (Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K., and Pease, L.R. (1989) Site-Directed Mutagenesis by Overlap Extension Using the Polymerase Chain-Reaction, Gene 77, 51-59.) removed. Specifically, first use the primer pair UC709-F / UC1179-R and UC1179-F / UC1695-R to amplify two DNA fragments, and then use the primer UC709-F / UC1746-R to splice the two DNA fragments by overlapping PCR Fragments, the spliced product was used as a large primer, and the plasmid pUC19 was used as a template to mutate one of the BmrI sites and three MlyI sites. The other two BmrI sites and MlyI sites located in the multiple cloning site sequence on the plasmid ...
Embodiment 2
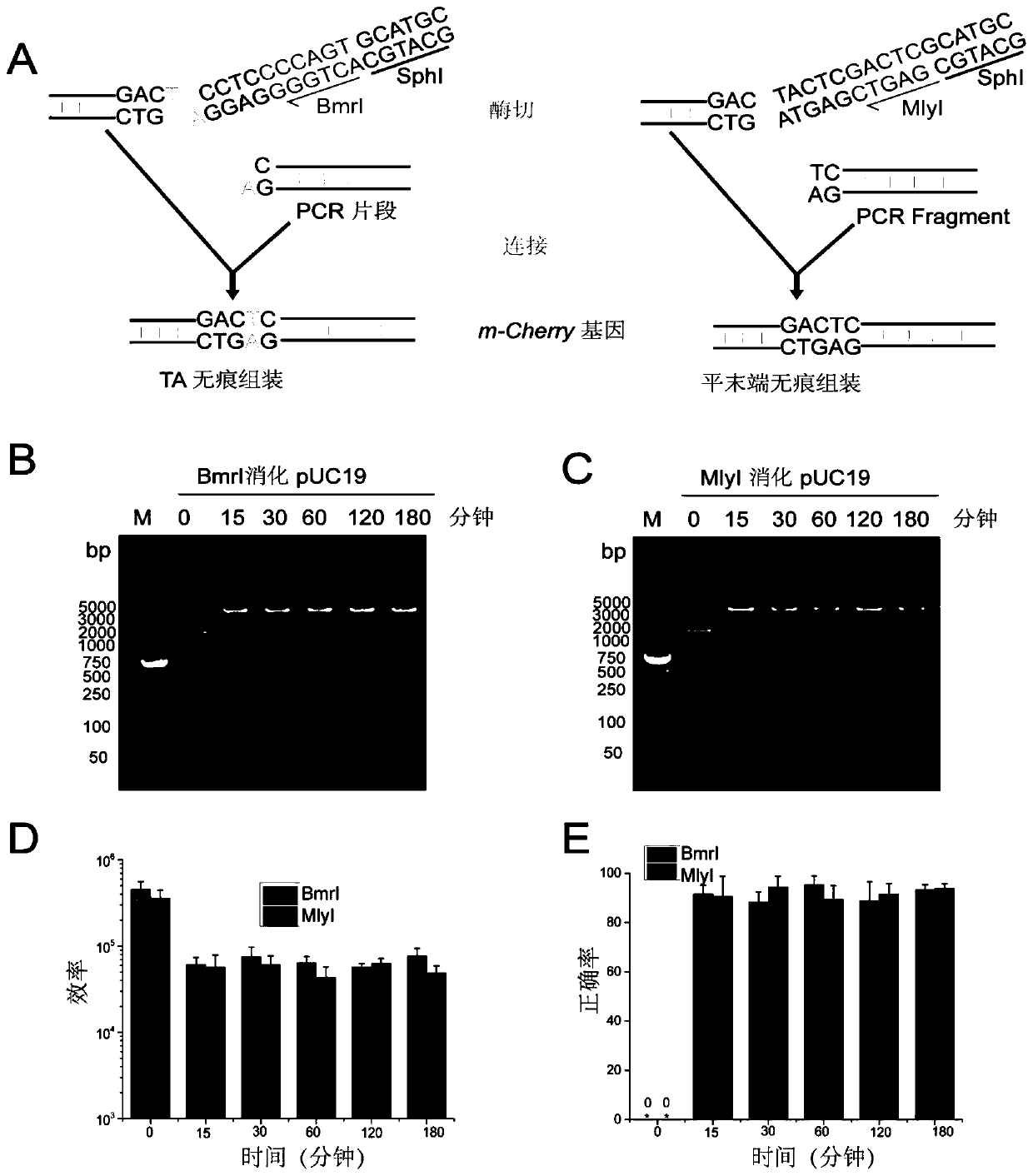
[0065] The reaction condition optimization of embodiment 2PS-Brick assembly
[0066] Use IIS-type restriction endonucleases BmrI and MlyI to single-enzyme pOB and pOM vectors respectively. The enzyme digestion reaction system is as described in Example 1. According to the reaction conditions of Time-Saver products, the reaction time is from 15 minutes to 180 minutes, and then electrophoresis Check cutting efficiency. Such as figure 2 As shown in BC, almost all vectors were completely cleaved within 15 min, yielding a single band of linearized size.
[0067] Cut the gel to recover the bands of pOB cut by BmrI and pOM cut by MlyI, and then digested with SphI for 15 to 180 minutes, inactivated at 65°C, recovered, ligated with the PCR products FB and FM cut by SphSphI respectively, and transformed into E. coli DH5α competent cells. The number of transformant units (colony-forming units (CFUs)) represents the DNA assembly efficiency. 20 clones were picked for sequencing, and t...
Embodiment 3
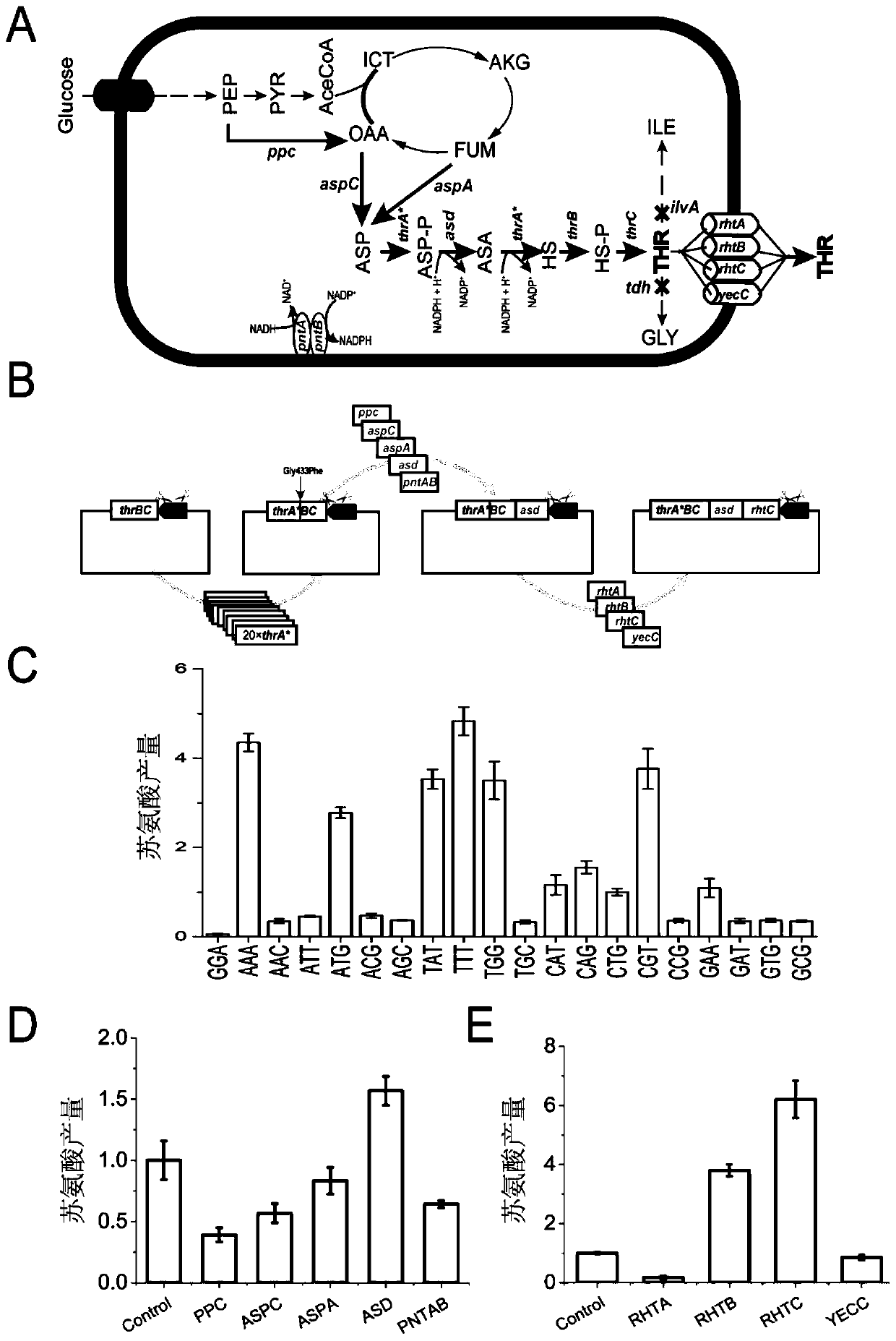
[0070] Example 3 Engineering Transformation of Threonine Metabolic Pathway Using PS-Brick Method
[0071] The PS-Brick assembly technology was used to carry out multiple rounds of iterative "design-build-test" cycles, and a threonine-producing engineered bacterium was constructed. Due to the complexity of the regulation and interaction of gene expression and signaling networks in organisms, and the lack of prior knowledge to predict how a DNA assembly introduced into a cell will function, it is necessary to test multiple versions of the construct to obtain optimal results. Assembly scheme. Multiple rounds of step-by-step "design-build-check" experiments require an iterative nature of the DNA assembly approach.
[0072] The metabolic engineering strategy for constructing threonine engineered bacteria usually includes the following steps: release the feedback inhibition of the threonine operon, enhance the threonine terminal synthesis pathway, remove the metabolic bottleneck, b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com