A kind of short-chain dehydrogenase and its application
A short-chain dehydrogenase and reaction technology, applied in the field of bioengineering, can solve the problem of low yield of ring-opening reaction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1 Cloning of short-chain dehydrogenase gene
[0061] According to the sequence of the Bacillus subtilis (Bacillus subtilis) short-chain dehydrogenase gene (NCBI accession number: WP_013058339) recorded by NCBI, the PCR primers were designed as follows:
[0062] BsSDR10 f:5ˊ-CGGGATCCGATGAAGTACACAGTTATTACAGGA-3ˊ;
[0063] BsSDR10r:5ˊ-CCGCTCGAGCATCTCTAGAATCGGATAGATTTGA-3ˊ.
[0064] The underlined part of the upstream primer is the BamHI restriction site, and the underlined part of the downstream primer is the XhoI restriction site.
[0065] PCR amplification was performed using the genomic DNA of Bacillus subtilis CGMCC 1.3358 as a template. The PCR system is as follows:
[0066]
[0067] Amplification program: 94°C for 10 min, 35 cycles of (94°C for 30s, 50°C for 30s, 72°C for 60s), 72°C for 10 min, and cooling to 4°C.
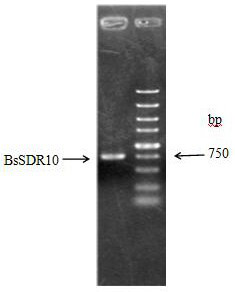
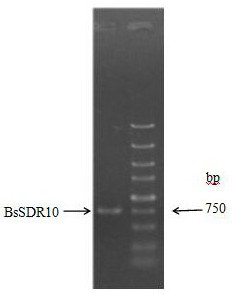
[0068] The PCR product was purified by agarose gel electrophoresis, and the target band of about 750bp was recovered using a DNA recovery ...
Embodiment 2
[0069] Example 2 Construction of recombinant expression vector plasmid and preparation of recombinant expression transformant
[0070] The DNA fragment of the short-chain dehydrogenase gene in Example 1 was double-digested with restriction enzymes BamHI and XhoI at 37°C for 12 h, the product was purified by agarose gel electrophoresis, and the target fragment was recovered using a DNA recovery kit. Under the action of T4 DNA ligase, the target fragment was ligated with the plasmid pET-28b-MBP which was also double digested with restriction enzymes BamHI and XhoI at 16°C overnight to obtain the recombinant expression transformant pET-28b -MBP-BsSDR10. Plasmid construction map such as Figure 5 shown.
[0071] The above recombinant expression vector plasmid pET-28b-MBP-BsSDR10 was transformed into Escherichia coli (E.coli) BL21 competent cells. Screen the positive recombinants on the plate, pick a single colony, and verify the positive clones by PCR. figure 2 shown.
Embodiment 3
[0072] Example 3 Expression of short-chain dehydrogenase BsSDR10
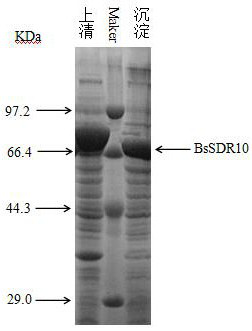
[0073] The recombinant Escherichia coli obtained in Example 2 was inoculated into the LB medium (PH 7.0) containing kanamycin, and incubated overnight at 37°C with shaking. In a 250ml Erlenmeyer flask of culture medium, place it in a shaker at 37°C and 180rpm for shaking culture. When the OD of the culture medium is 600 When reaching 0.6, IPTG with a final concentration of 0.25 mM was added as an inducer, induced at 20 °C for 12 h, the culture medium was centrifuged, cells were resuspended with 3 ml of phosphate buffer solution (pH 6.0), and transferred to EP tubes. The supernatant was collected by centrifugation at low temperature (4°C), which was the crude enzyme solution of recombinant short-chain dehydrogenase. The crude enzyme solution and the precipitate were analyzed by polyacrylamide gel electrophoresis, such as image 3 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com