Novel coronavirus T cell epitope peptide, pMHC and preparation and application of novel coronavirus T cell epitope peptide
A coronavirus, cell antigen technology, applied in the direction of viral peptides, viruses, viruses/phages, etc., can solve the problem that there is no T cell antigen epitope report or suggestion.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1 Prediction of New Coronavirus HLA-A2 Restricted Epitopes
[0064] 1. Experimental method
[0065] The spike(S) and membrane of 2019-nCoV (MN908947.3), SARS China isolate (DQ182595.1), MERS Riyadh isolate (KF600612.1) and 1997 US common coronavirus isolate (KF530099.1) were taken (M) and nucleocapsid (N) protein sequences, compared with Clustal Omega. Epitopes inconsistent with SARS, MERS and common coronavirus sequences were selected for follow-up research. Simultaneous selection of known HLA-A2 restricted CD8 + T cell epitopes served as positive controls.
[0066] 2. Experimental results
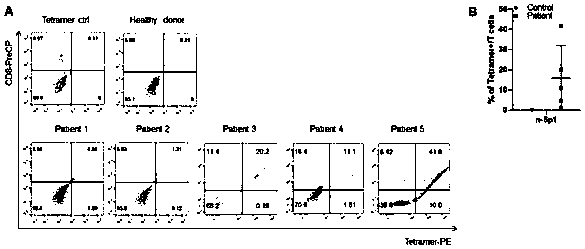
[0067] A large number of high-efficiency and specific T cell antigen epitope candidates were screened.
Embodiment 2
[0068] Example 2 Identification of New Coronavirus HLA-A2 Restricted Epitopes
[0069] 1. Experimental method
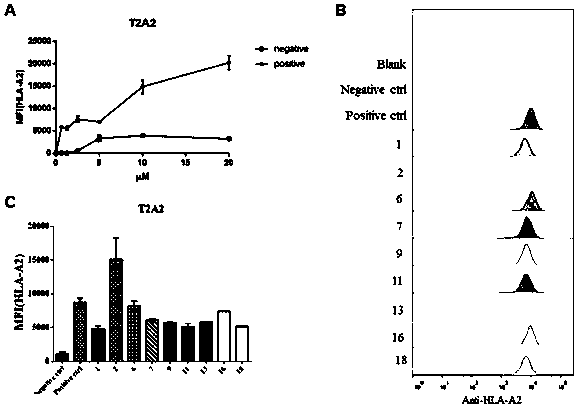
[0070] The peptides predicted in Example 1 were artificially synthesized and prepared in different concentrations, namely 0.625 μM, 1.25 μM, 2.5 μM, 5 μM, 10 μM, and 20 μM. Take T2-A2 cells in the logarithmic growth state and plant them into 96-well plates, 10 per well 5 , distribute and configure blank wells, negative control peptide (GLQRLGYVL, derived from Zika virus gene code), positive control peptide (influenza A M1 polypeptide, GILGFVFTL) and each synthetic antigen polypeptide, each group has 3 duplicate wells, and the final volume is 200 μL. After incubation at 37°C for 4 hours, centrifuge and wash twice, label with FITC anti-human HLA-A2 (β2m) antibody, incubate at 4°C for 30 minutes in the dark, and detect with flow cytometry. The experiment was carried out 3 times.
[0071] 2. Experimental results
[0072] The result is as figure 1 As shown, the resul...
Embodiment 3
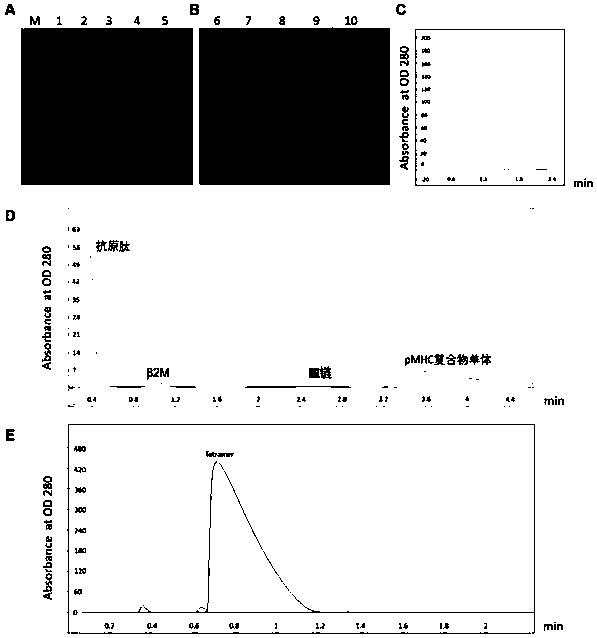
[0075] Example 3 Preparation of pMHC complex of novel coronavirus antigen epitope
[0076] 1. Preparation of HLA-A2 heavy chain and light chain
[0077] 1. Experimental method
[0078] Recombinant construction of genetic engineering technology The heavy chain HLA-A2 (NCBI No. U02935.2) and the light chain β2m (NCBI No. AY187687.1) two target genes were respectively connected to the prokaryotic expression vector pTXB1 through SpeI and NdeI to obtain pTXB1-HLA-A2h and pTXB1-β2m prokaryotic expression plasmids. The plasmid was transformed into BL21 Escherichia coli by heat shock method (42°C for 90 seconds), and sequenced and analyzed. The bacteria liquids successfully identified by sequencing were stored in 30% glycerol at -20°C. Inoculate 10 μL of glycerol bacteria into 4 mL of ampicillin-containing LB medium, culture at 37°C 280 rpm for 16-18 hours, then expand the culture to 400 ml, culture at 37°C 280 rpm for 4 hours, induce expression with 0.1mM IPTG at 24°C 280 rpm for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com