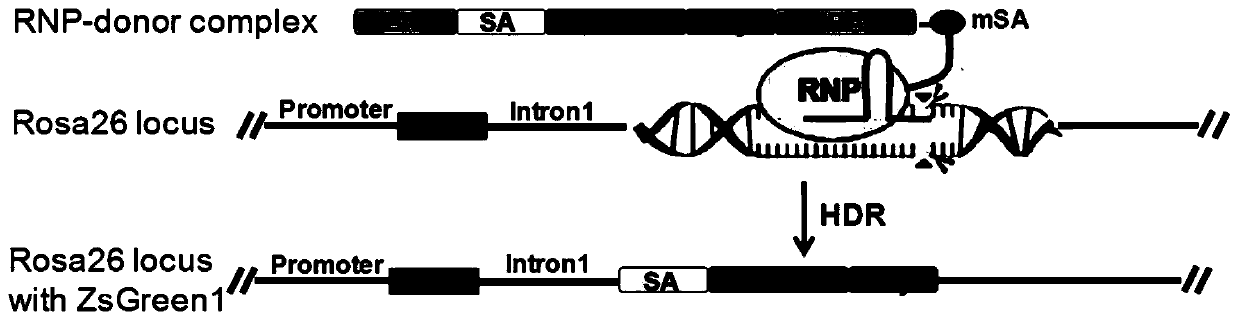
Efficient clustered regularly interspaced short palindromic repeats ribonucleoprotein (CRISPR RNP) and donor DNA co-location mediated gene insertion or replacement method and application thereof
A donor and gene technology, applied in the field of genetic engineering, can solve problems affecting the accuracy and efficiency of gene insertion or replacement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
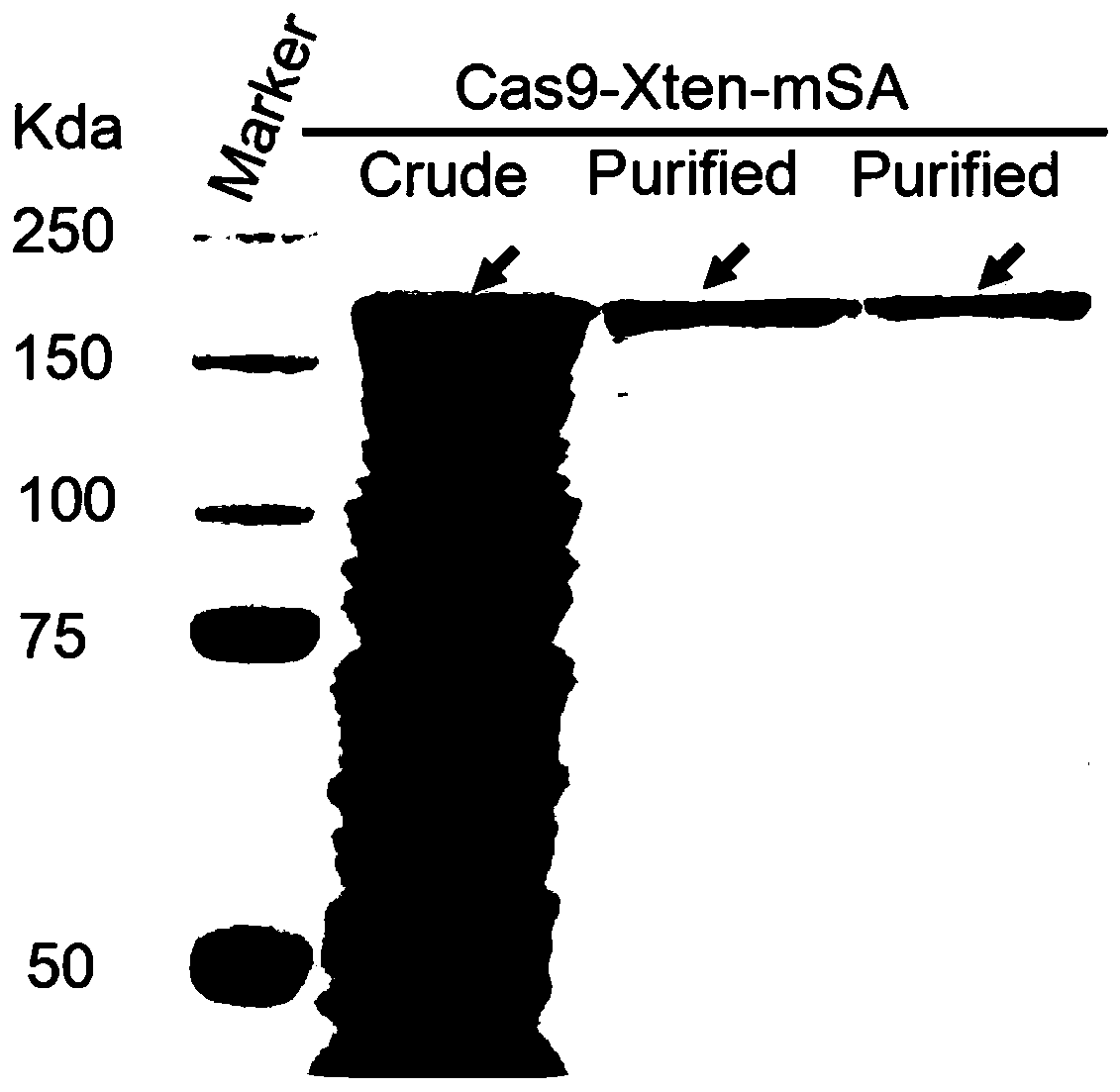
[0059] (1) Preparation of Cas9-Xten-mSA protein
[0060] a. Construction of expression plasmid pET-Cas9-Xten-mSA
[0061] Synthesize the DNA (SEQ ID NO.5) sequence encoding Xten-mSA, and clone it into the pET-NLS-Cas9-NLS-6xHis vector by enzyme digestion and ligation to obtain the expression plasmid pET-Cas9-Xten-mSA; the specific steps are as follows :
[0062] The plasmid pET-NLS-Cas9-6xHis (Addgene) was purchased through Beijing Zhongyuan Company. Using this plasmid as a template, primer 5'-CGCTAGAGCTCCCGCTGCTTTTAAATATTTTG-3'(SEQ ID NO.6) and primer 5'-GCAGCTTCAACTTTTTCTTTTTGTCACTCCTAGCTGACTCAAATC-3'( SEQ ID NO.7) was amplified to obtain fragment I; using primer 5'-TGACAAAAAGAAAAAGTTGAAGCTGCATCACCACCATCACTAATG-3' (SEQ ID NO.8) and primer 5'-GCAGCCTAGGTTAATTAAGCTGCGCTAG-3' (SEQ ID NO.9) to amplify, Fragment II is obtained. Using the Overlap Extension PCR method, the mixture of fragments I and II in equimolar ratio was used as a template, and the above primers (primer SEQ ...
Embodiment 2
[0080] Example 2 Observation and Flow Cytometry Analysis of ZsGreen1 Gene Expression in Nuclear Transformed Cell Population
[0081] (1) After 24 hours of nuclear transformation in step (6) of Example 1, the expression of the ZsGreen1 gene was observed under a Zeiss Axio Observer A1 fluorescence inverted microscope, and the CRISPR RNP-donor DNA complex (sgRNA-Cas9-Xten-mSA- After ZsGreen1) nuclear transformation, about 10% of the cells showed fluorescence ( Figure 7 A); as a control, few fluorescent cells appeared after nuclear transformation of the CRISPR RNP-donor DNA hybrid (sgRNA-Cas9+ZsGreen1) ( Figure 7 B). Since the ZsGreen1 gene has no promoter when designing the donor DNA, it can only be expressed in the first intron of the Rosa26 gene through homologous recombination and depends on the promoter of the Rosa26 gene, so the fluorescent cells are correctly inserted into the ZsGreen1 gene cells at the target site. Wherein, the nuclear transformation of CRISPR RNP and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com