Primer set, probe set and kit for detecting COVID-19 virus, and application of kit
A COVID-19 and kit technology, applied in the field of microbial detection, can solve the problems of high operator skill requirements, large sequencing data volume, high sample requirements, etc., achieve large market prospects, wide application scenarios, and improve sensitivity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] (1) Preparation of luminescent label-No. 1 probe complex:
[0070] 1) Select acridinium ester as a luminescent marker, and the acridinium ester solution is prepared by the following method:
[0071] Dissolve 5 mg of acridinium ester with 1.58 mL of DMSO to obtain a mother solution of acridinium ester with a concentration of 3.164 mg / mL, which is set aside. The acridinium ester mother liquor was diluted 10 times with DMSO to obtain an acridinium ester solution with a concentration of 0.3164 mg / mL.
[0072] 2) Preparation of No.1 probe solution:
[0073] Take probe No. 1 bound with biotin and dissolve in 0.1M NaHCO 3 , and the volume was adjusted to 90 μl to obtain the No. 1 probe solution.
[0074] 3) Mix the acridinium ester solution in step 1) with the No. 1 probe solution in step 2), control the mass ratio of the acridinium ester to the No. 1 probe to be (4-16): 1, and then place the mixed solution at room temperature The reaction was carried out for 5 hours, and ...
Embodiment 2
[0079] A kit, comprising: PCR amplification reaction system, No. 1 probe, No. 2 probe, acridinium ester solution, magnetic beads, cleaning solution, and excitation solution.
[0080] The No.1 probe is: 5’-ctcgttgact ttcaggttac tatagcagag at-Biotin-3’
[0081] The second probe is: 5'-NH 2 -ctaactgaga ataaatattc tcaattagat ga-3';
[0082] Among them, the No. 1 probe and the acridinium ester solution can be prepared into a luminescent marker-No. 1 probe complex by the method described in Example 1; the No. 2 probe and the magnetic beads can be formed by the method described in Example 1. Magnetic Beads - No. 2 Probe Complex.
[0083] The PCR amplification reaction system includes: 1.0 μL 10×Buffer, 0.2 μL dNTPs, 1.0 μL 2U / μL Taq enzyme, 0.1 μL 1.25 μmol / L upstream primer, 1.0 μL 1.25 μmol / L downstream primer, template, etc., ddH 2 O to make up to 10 μL;
[0084] The upstream primer is: 5'-at ctcgttgact ttcaggttac-3'
[0085] The downstream primer is: Phos phosphate group-5'-...
Embodiment 3
[0088] Adopt the kit detection COVID-19 virus of embodiment 2, comprise the following steps:
[0089] (1) Asymmetric PCR amplification
[0090] Using the serum of a patient with COVID-19 virus infection confirmed by sequencing as a sample, RNA was extracted, and asymmetric PCR amplification was performed after reverse transcription to obtain the target single-stranded DNA.
[0091] Among them, the PCR amplification program is: 95°C pre-denaturation for 5 minutes; 95°C denaturation for 15s, 55°C annealing for 30s, 72°C extension for 60s, 40 cycles, and purification to obtain PCR amplification products.
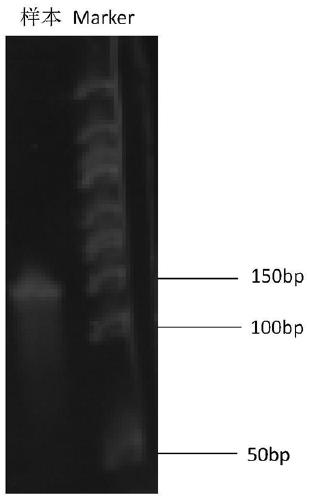
[0092] Adopt 15% agarose electrophoresis to detect PCR product, the result is as follows figure 1 As shown, an obvious band appeared at about 150bp.
[0093] (2) One hybridization reaction
[0094] According to the method described in Example 1, the luminescent marker-No. 1 probe complex was prepared, wherein the mass ratio of the luminescent marker acridinium ester to the N...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Particle size | aaaaa | aaaaa |
| Density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com