Method for rapidly detecting SARS-CoV-2 based on DNA nano-scaffold
A nano-scaffold, sars-cov-2 technology, applied in the field of genes, achieves the effects of programmable sequence, simple operation and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
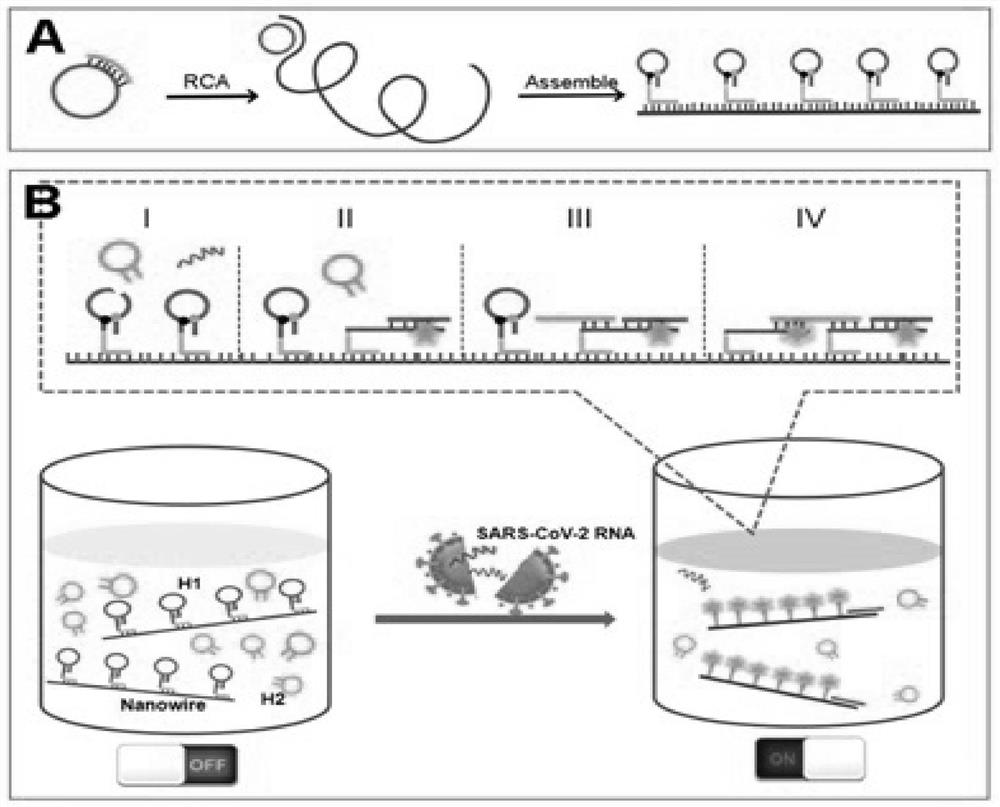
[0039] A method for rapidly detecting SARS-CoV-2 based on DNA nano-stents of the present invention comprises the following steps:
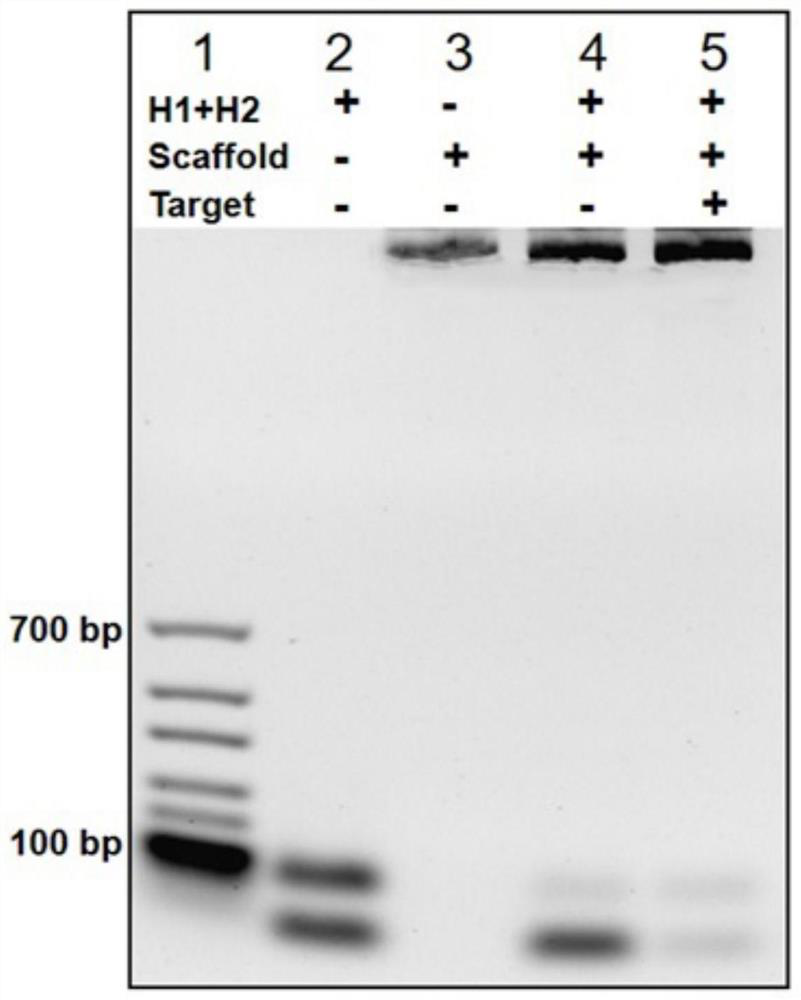
[0040] (1) A DNA nanoscaffold was constructed by DNA strand self-assembly and self-quenching probe H1, and the target RNA triggered the free hairpin probe H 2 and hairpin probe H 1 Rapid hybridization along the nanoscaffold enables signal amplification.
Embodiment 2
[0042] A method for rapidly detecting SARS-CoV-2 based on DNA nano-stents of the present invention comprises the following steps:
[0043] (1) by DNA hairpin probe H 1 Self-assembly with DNA nanowires to construct nanoscaffolds, in which RCA reaction by Pre-cirDNA and Primer produced DNA nanowires containing repeat sequence fragments, and hairpins were labeled with 5-carboxyfluorescein FAM dye and its fluorescent quencher BHQ1 both ends of probe H1 to form a self-quenching probe;
[0044] (2) When the target RNA is present, based on the H in the nanoscaffold 1Encoding design, which will hybridize with one H1 in the nanoscaffold, so that the hairpin probe H1 hairpin is opened and the fluorescence is restored, which in turn triggers the cascade of hairpin probe H2 and hairpin probe H1 along the DNA nanoscaffold hybridization; the Pre-cirDNA has the nucleotide sequence of SEQ ID No.4. The Primer has the nucleotide sequence of SEQ ID No.5. The target RNA is the conserved regio...
Embodiment 3
[0057] Optimize experimental conditions
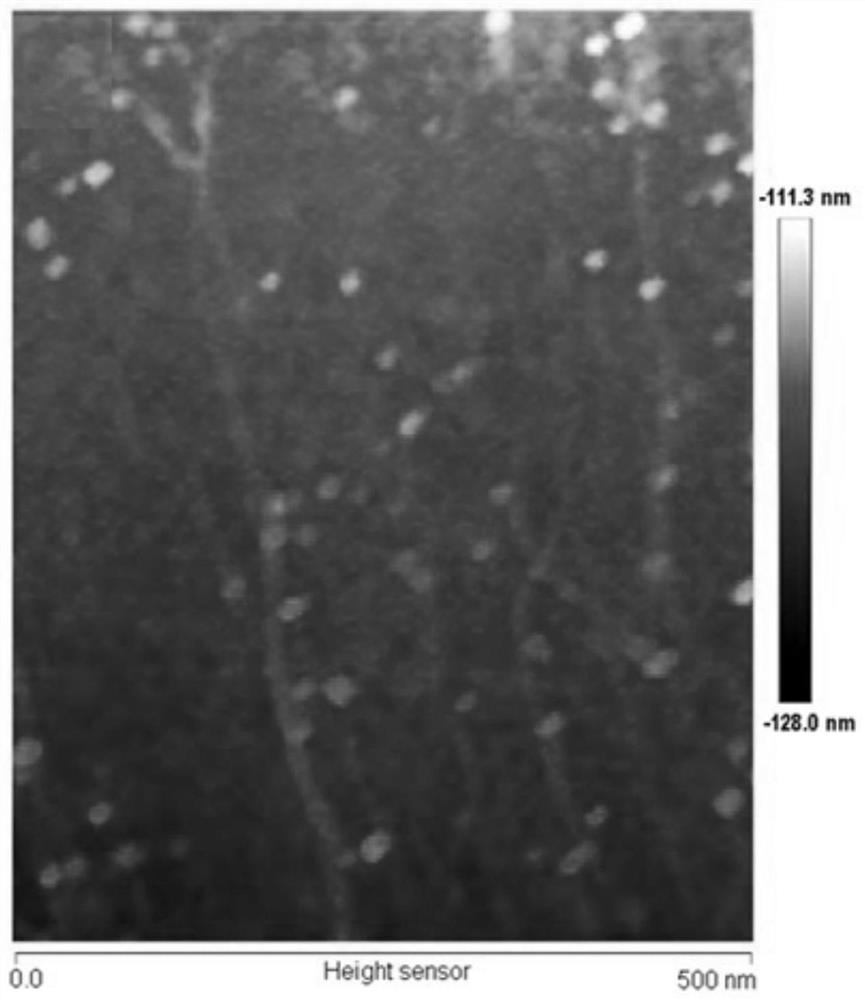
[0058] In order to obtain better detection efficiency, we optimized some key factors in the experiment process. The first is the effect on the background signal of the length (foothold) at which H1 hybridizes to the DNA nanowire. The present invention optimizes the distance of the H1 probe on the nanowire, from Figure 5 It can be seen that there is the strongest fluorescent signal at 25 base pairs. In addition, the time of the RCA reaction affects the length of the generated DNA scaffold, which in turn changes the number of assembled H1 probes on the scaffold, ultimately affecting the sensitivity of the assay. like Image 6 As shown, the signal intensity increases with the prolongation of the RCA time from 0 min to 30 min, and decreases with the prolongation of the RCA time between 40 min and 100 min. Therefore, we choose 30 minutes as the optimal RCA response time. Finally, we explored the reproducibility of target RNA recogniti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com