HIV-1 viral load real-time fluorescence quantitative PCR detection specific primer pair and kit
A real-time fluorescence quantification, HIV-1 technology, applied in microbial determination/inspection, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problem that primers are difficult to be widely used and unsuitable for detection of HIV-1 infected persons. , to achieve the effect of shortening detection time, accurate viral load level, and reducing pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
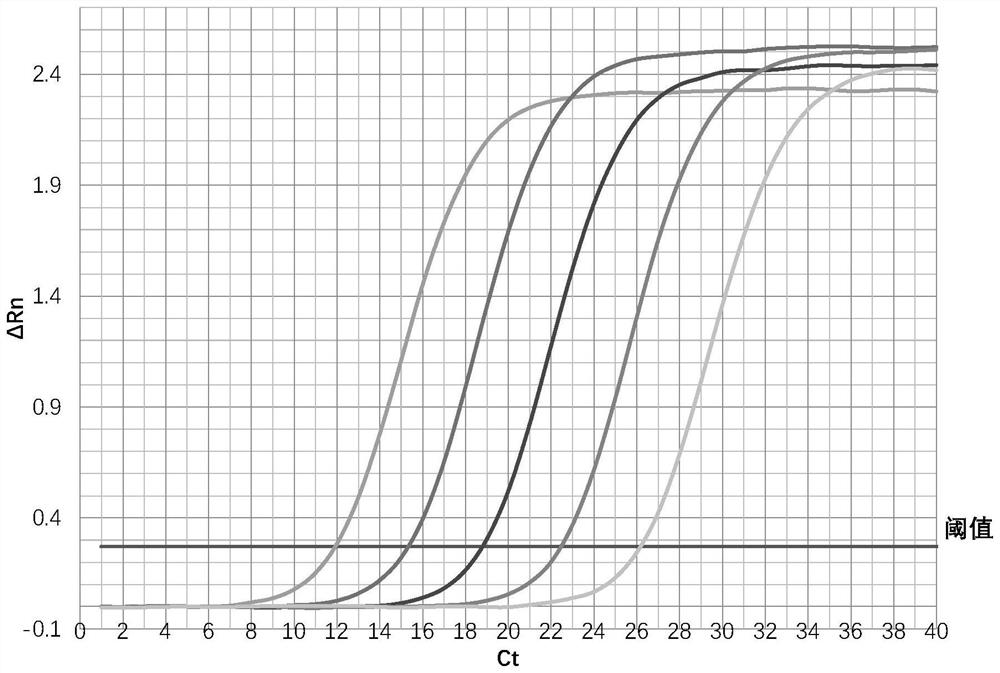
[0055] The RNA extracted from 16 HIV-1 positive serum samples was detected by using the human immunodeficiency virus type Ⅰ one-step fluorescent quantitative PCR detection kit.
[0056] Real-time fluorescence quantitative PCR was performed using Applied Biosystems QuantStudio5 Real-Time PCR Systems, and the fluorescence signal of the FAM channel was collected at 72°C, and the melting curve step was performed after the cycle process was completed.
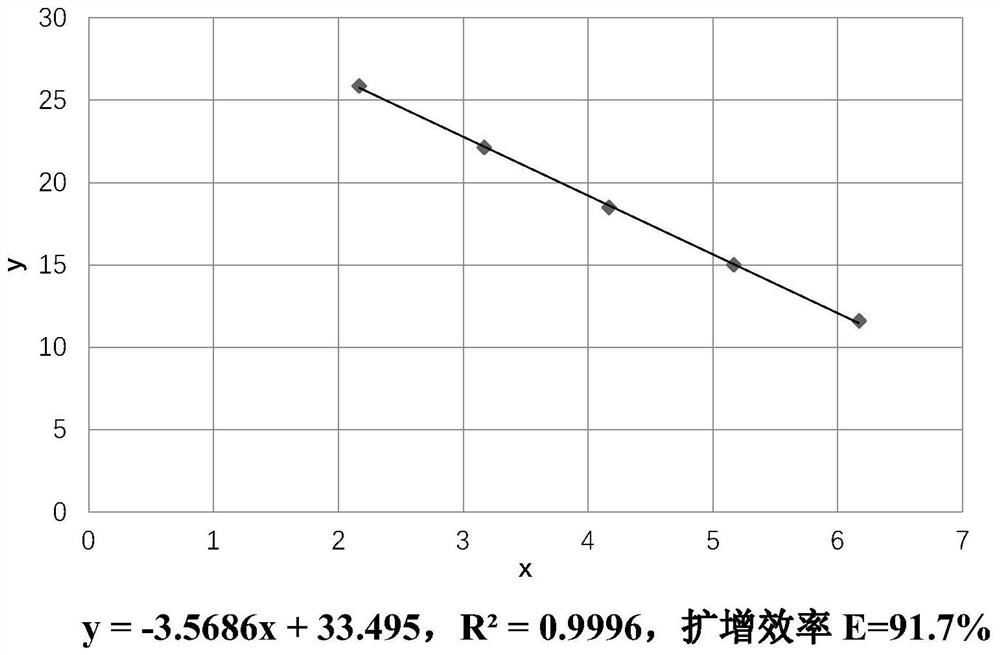
[0057] After the RT-PCR program of the instrument is completed, save the results and analyze the data according to the requirements of the instrument and software. Taking the fluorescence value higher than the sample noise line and the negative control as the detection threshold, the analysis software automatically combined the two standard curves to calculate the HIV-1 RNA content C (copies / mL) of each sample extract, and took the average as the final result. Then analyze the melting curve to ensure that there is no primer-dimer in...
Embodiment 2
[0065] Embodiment 2, to the detection result of various subtype HIV-1
[0066] The samples were subjected to first-generation sequencing and the resulting sequences were submitted to the Stanford University HIV Drug Resistance Database (https: / / hivdb.stanford.edu / hivdb) for subtype analysis. Afterwards, the viral load of each sample was detected using the kit provided by the present invention (the method and procedure are the same as in Example 1), and the results are as follows.
[0067] (1) Sample number R16, sequencing result (SEQ ID No: 5).
[0068] HIV-1 subtype is B; amplification result; Ct=26.681
[0069] (2) Sample number T1516, sequencing result (SEQ ID No: 6).
[0070] HIV-1 subtype is CRF01_AE; amplification result: Ct=23.259
[0071] (3) Sample number: Y4, sequencing result (SEQ ID No: 7).
[0072] HIV-1 subtype is CRF07_BC; amplification result: Ct=27.806
[0073] (4) Sample number: Y10, sequencing result (SEQ ID No: 8).
[0074] HIV-1 subtype is B+C; ampli...
Embodiment 3
[0076] Embodiment 3, sensitivity and stability verification test:
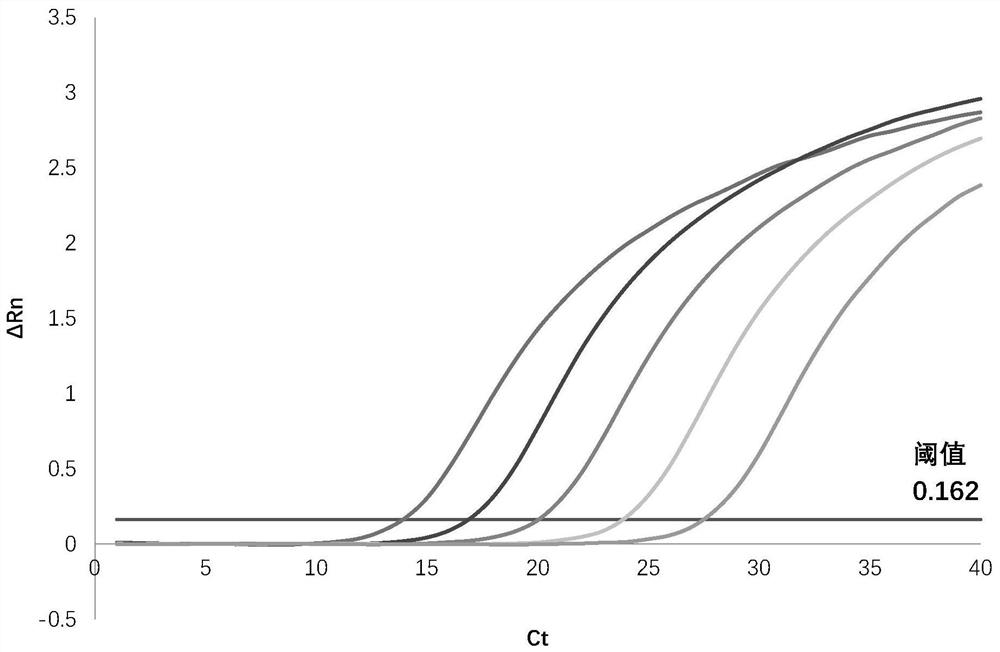
[0077] 1. Sensitivity verification test: The template DNA was serially diluted with a 10-fold gradient, starting from 5.92×10 3 ~5.92×10 9 In the copies / mL interval, 5 μL of each order of magnitude dilution was taken as the amplification template. The test conditions are the same as in Example 1. The result shows: obvious amplification curve can be seen at Ct=30 place (see Image 6 ), demonstrating that the lowest dilution that can be detected is 3 copies / mL (approximately equivalent to the minimum detection limit of 30 DNA copies per reaction), with good sensitivity, each reaction can detect a minimum of 30 copies of DNA.
[0078] 2. Stability verification test: Three sets of primers and standard products provided by the present invention are divided into three groups at one time, and stored at -20°C. On the 0th, 3rd, and 6th day, carry out RT-PCR amplification test respectively (test method and program a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com