Specific SS-COI primer pair of hypothenemus hampei Ferrari, identification kit and rapid identification method
A technology of coffee cherries and primer pairs, which is applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as money-consuming, time-consuming and labor-intensive, and unfavorable insect invasion and diffusion dynamics.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1: Extraction of insect genome DNA
[0047] Genomic DNA was extracted using the Ezup Column Animal Genomic DNA Extraction Kit from Sangon Bioengineering Co., Ltd. The specific operation steps are as follows:
[0048] (1) Clean the single-headed beetle with absolute ethanol for 5 seconds, then wash it with sterile water for 5 times, and place it on a sterilized filter paper to dry;
[0049] (2) Put the dried beetle into a 1.5mL centrifuge tube, add 180μL BufferACL, grind the sample thoroughly with an electric grinder, then add 20μL Proteinnase K solution, shake and mix well, and bathe in 56℃ for 1 hour until the cells are completely lysed ;
[0050] (3) Add 200μL Buffer CL and mix thoroughly by inversion;
[0051] (4) Add 200 μL of absolute ethanol and mix thoroughly by inversion;
[0052] (5) Put the adsorption column into the collection tube, add all the solution and translucent fibrous suspension into the adsorption column with a pipette, let it stand for 2...
Embodiment 2
[0056] Embodiment 2: PCR amplification of bark beetle insect COI gene
[0057] According to the report of Jordal et al. (2011), S1718 / A2237 was selected as the universal primer for bark beetle COI gene amplification:
[0058] S1718: 5'-GGAGGATTTGGAAATTGATTAGTTCC-3'
[0059] A2237: 5'-CCGAATGCTTCTTTTTTTACCTCTTTCTTG-3'
[0060] Use Tiangen 2×Taq PCR MasterMix II kit for PCR amplification reaction, the total reaction system is 25 μL, including:
[0061]
[0062] Amplification conditions: pre-denaturation at 94°C for 4 min; denaturation at 94°C for 30 s, annealing at 46°C for 50 s, extension at 72°C for 1 min, 40 cycles; extension at 72°C for 10 min.
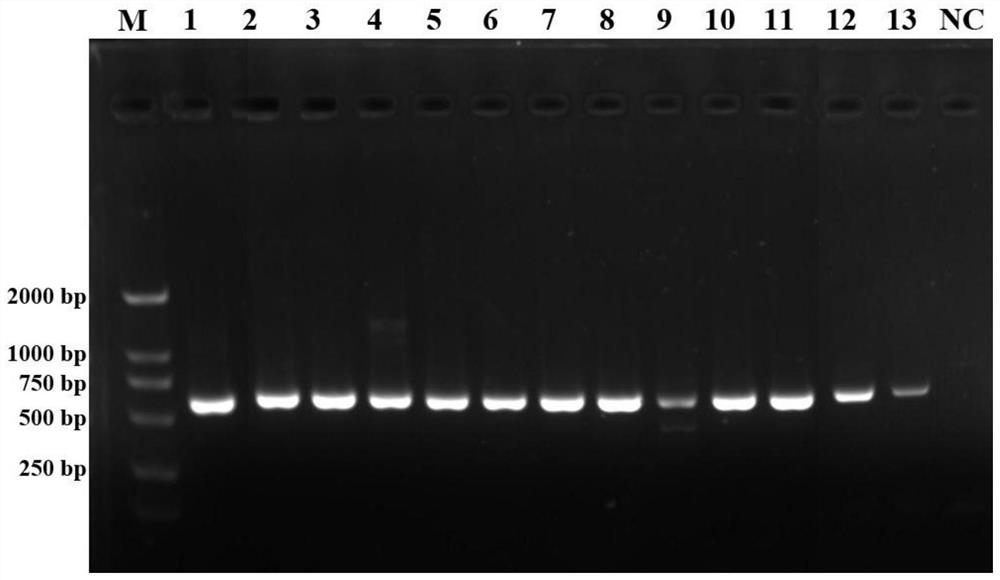
[0063] Take 5 μL of the PCR product, and separate it by electrophoresis on a 1.2% agarose gel containing Gel stain staining agent for 20 min (140V), detect the result with a gel imaging system, and take pictures.
[0064] The results showed that the COI gene fragments of 13 bark beetle insects including the coffee berry beetle...
Embodiment 3
[0065] Example 3: Design of specific primers for the coffee berry beetle COI gene
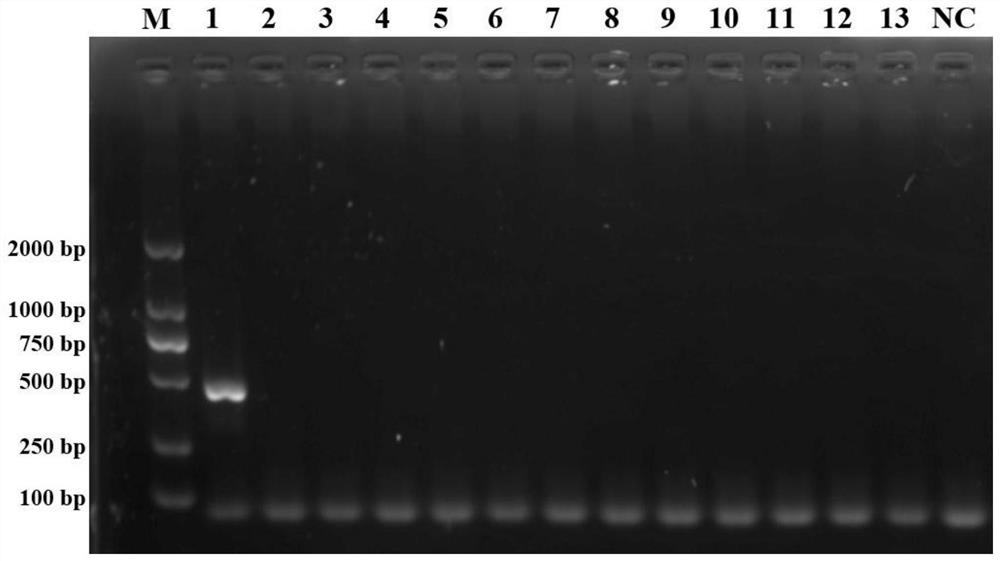
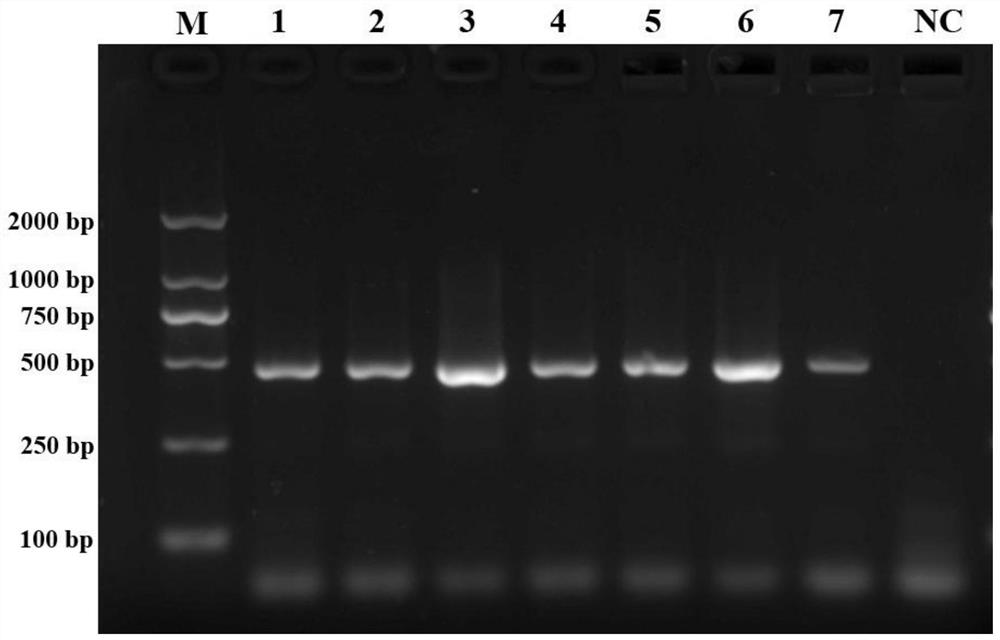
[0066] The COI sequences of 13 bark beetle insects obtained in this study (including coffee berry bark beetle) were verified by Blast sequence alignment in NCBI. The results showed that the COI fragments of 13 bark beetle insects were successfully amplified using the universal primer S1718 / A2237. The 13 COIs were then subjected to multiple sequence alignment analysis, and a pair of COI-specific primers (SEQ ID NO: 1, SEQ ID NO: 2) for the coffee berry beetle were designed using the PrimerSelect software in DNASTAR:
[0067] HHTYF1: AAGGTGTTGATATAGGATTGGGTCC
[0068] HHTYR3: CCACTAATACTAGGCGCACCTG
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com