Leucine dehydrogenase mutant and construction method and application thereof
A technology of leucine dehydrogenase and construction method, which is applied in the field of leucine dehydrogenase mutants and its construction, and can solve problems such as inability to catalyze chiral amino acids
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Preparation of Leucine Dehydrogenase Mutants
[0050] The leucine dehydrogenase derived from Bacillus pumilus was used as the wild type, that is, WT EsiLeuDH.
[0051] (1) Ligate WT EsiLeuDH DNA with plasmid pET28a to obtain a recombinant vector.
[0052] (2) Obtain mutant K89T:
[0053] Using the DNA of the recombinant vector as a template, K89 saturated-IR (the nucleotide sequence is shown in SEQ ID NO.2, specifically: GATGATGACCGCNNNCCCG) and K89 saturated-IF (the sequence is shown in SEQ ID NO.3, specifically: : GCGGGNNNGCGGTCATCAT) was used as a primer to carry out PCR site-directed saturation mutation to obtain a PCR product. Among them, the PCR reaction system is as follows: 1 μL of WT EsiLeuDH DNA at 15 ng / μL, 1 μL of mutant primers IF and IR (10 μM), 15 μL of 2X Trans startfastPfu, and ddH2O supplementation system to 30 μL. PCR reaction program: pre-denaturation at 94°C for 4 min; denaturation at 94°C for 20 s, annealing at 60°C for 20 s, extension at 72°C f...
Embodiment 2
[0061] Expression and purification of mutants
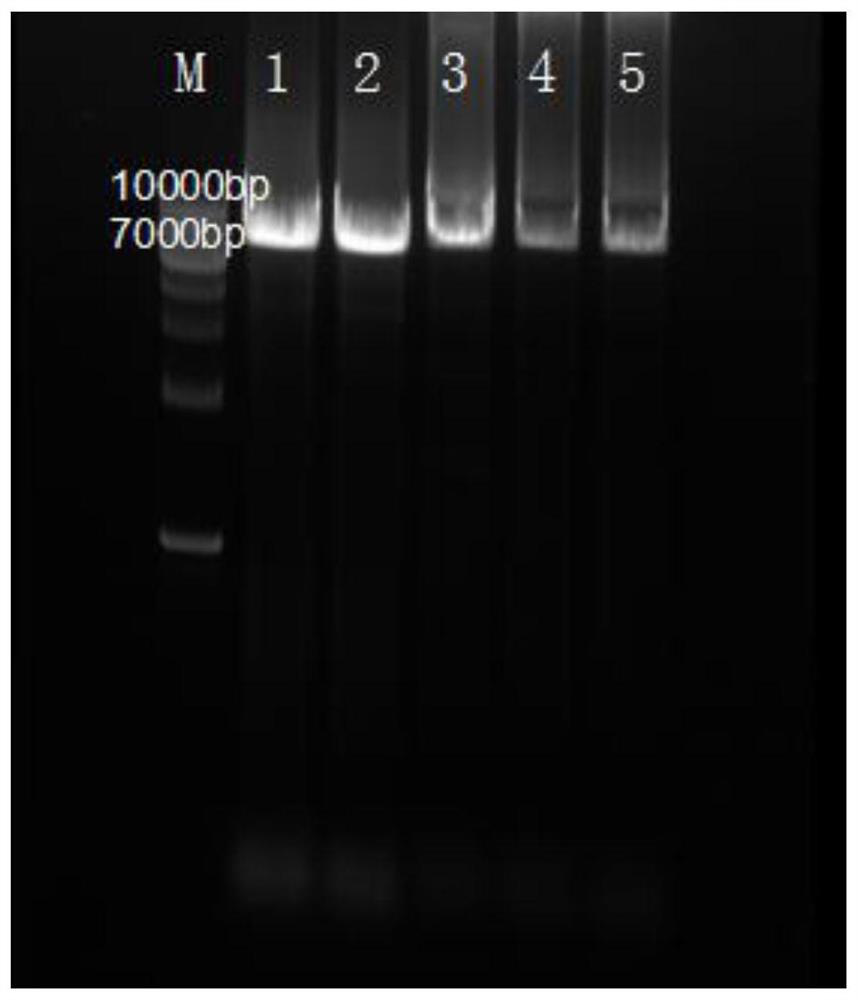
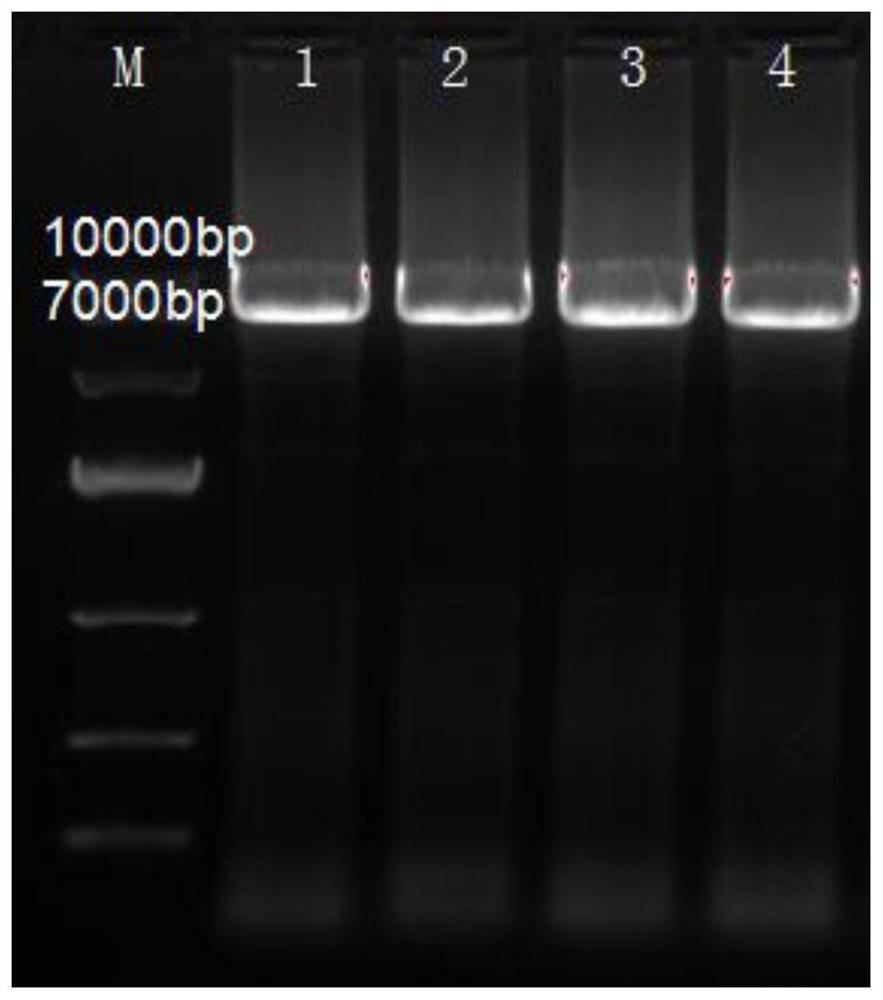
[0062] The PCR product obtained in Example 1 was digested with DPN1 enzyme at 37° C. for 2 h. After the digestion treatment, the product was directly transformed into Escherichia coli BL21 (DE3) competent cells by heat shock method to obtain recombinant strains. Spread the obtained recombinant strains on a plate containing Kana antibiotics, and place the plate in a 37°C incubator for 12 hours. After the colonies grow, pick the colonies for PCR colony verification and mutation library screening, followed by liquid culture. A portion of the bacterial culture was sent for sequencing. PCR colony verification and sequencing results showed that mutant A122G and K89T genes had been constructed and successfully introduced into Escherichia coli BL21(DE3).
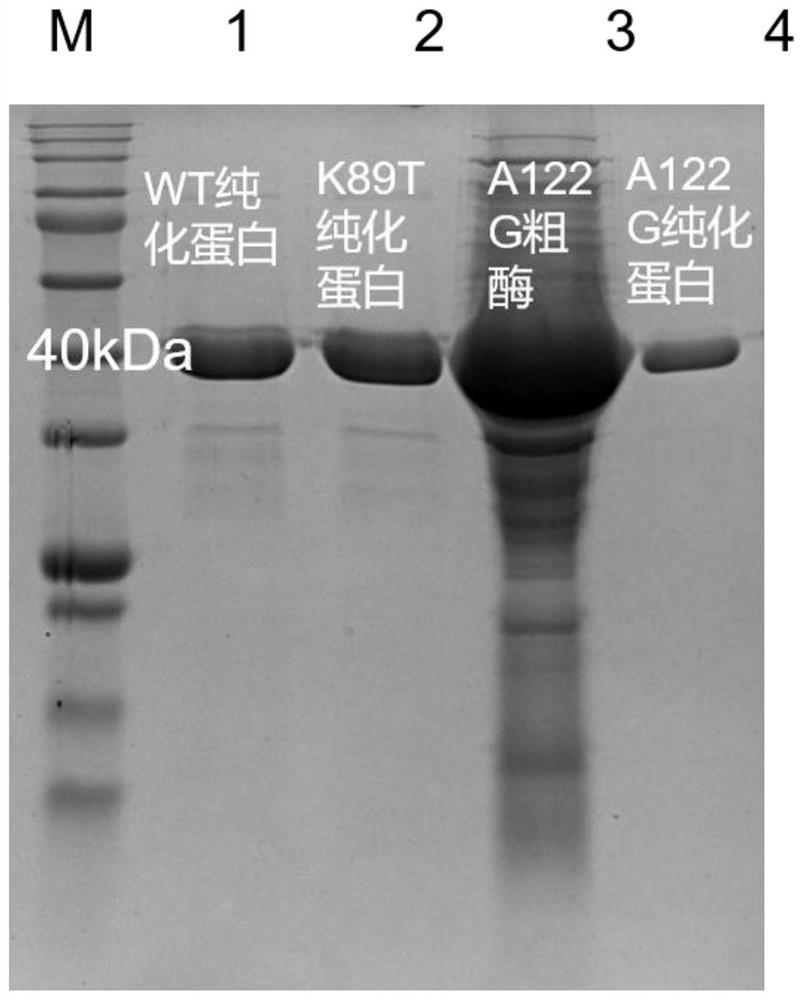
[0063] The obtained recombinant strain was expanded and cultivated, and the expression of the mutant protein was induced by the chemical reagent IPTG induction method. After the indu...
Embodiment 3
[0066] Enzyme activity assay of leucine dehydrogenase
[0067] The WT purified protein, K89T purified protein and A122G purified protein obtained in Example 2 were subjected to substrate spectrum determination. The substrate spectrum determination system: measured by a microplate reader, the total volume of the reaction system was 220 μL, and the optical path was 0.5 cm. After proper dilution of amino acid dehydrogenase, take 10 μL, 4mM reaction substrate (any one of the 20 kinds of amino acids in the laboratory) 50 μL, 20mM coenzyme NAD+11 μL, 0.2M, pH9.5 glycine-sodium hydroxide 149 μL of buffer solution; measure the increase in NADH concentration at a wavelength of 340 nm, and the enzyme activity of one unit of leucine dehydrogenase is defined as the amount of enzyme required to reduce 1 μmol NAD+ within 1 min.
[0068] Enzyme activity assay principle:
[0069] Enzyme activity (U / mL) = ΔA / Δt×Vt / (Vs×L×ε);
[0070] Specific enzyme activity (U / mg) = enzyme activity (U / mL) / ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com