Application of vitreoscilla hemoglobin in improving expression quantity of cephalosporin C acylase
A technology of cephalosporin and Vibrio vitreous, applied in application, enzyme, bacteria and other directions, can solve the problems of difficult reaction control, unfriendly chemical deacylation method, harsh reaction conditions, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] [Example 1] Experimental material
[0036] Strains and plasmids
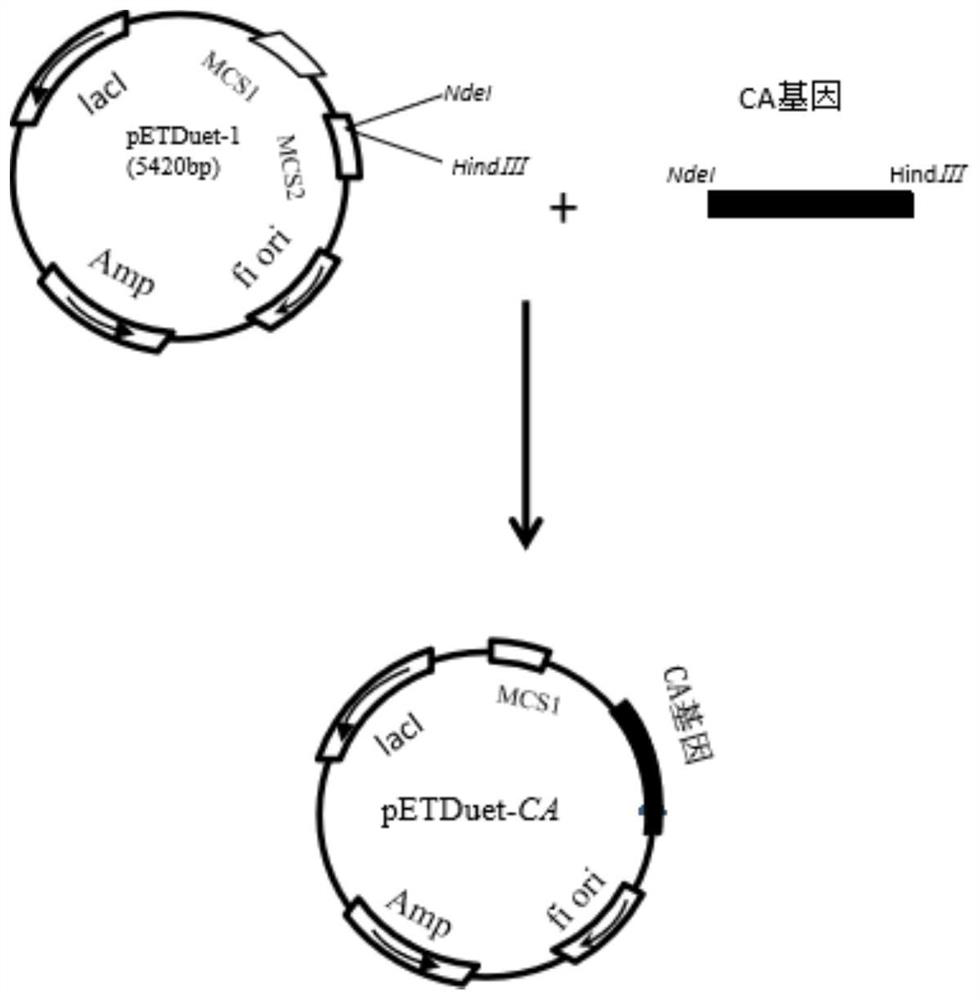
[0037] Escherichia coli E.coli BL21(DE3) and Escherichia coli DH5α were purchased from Shenzhen Kangti Life Technology Co., Ltd. Plasmids pETDuet-1, pACYCDuet-1, pRSFDuet-1 were purchased from Addgene.
[0038] experimental method:
[0039] 1. P1 phage transduction recipient bacteria
[0040] (1) Lysis of the donor bacterium JM1087 (JM1087 genotype: BW25113 ptsG::kan, phenotype: Kan+, the ptsG gene of the donor bacterium has been knocked out, and the corresponding position is replaced by the kanamycin resistance gene, which with FRT sequences at both ends)
[0041] ①Cultivate donor bacteria JM1089 in 5 mL of LB until OD600=0.1-0.2, add 50 μL of 1M CaCl2, and mix well.
[0042] ② Add 10-100 μL of wild-type P1 phage, and incubate on a shaker at 37°C for at least 3 hours until the cells are lysed.
[0043] ③ Add 1 to 3 drops of chloroform to each ml of culture medium and shake to kill the cells.
[00...
Embodiment 2
[0104] [Example 2] Experimental method
[0105] Cloning and Sequence Analysis of Cephalosporin C Acylase Gene and Vitiligo hyaline Hemoglobin Gene
[0106] The cephalosporin C acylase gene is re-synthesized according to the codon recognition characteristics of Escherichia coli to optimize the GC content, as shown in SEQ ID NO.9, which is more conducive to its expression in Escherichia coli.
[0107] Strain recovery
[0108] 1. Take out the strains stored at -80°C and dissolve them on ice.
[0109] 2. Use an inoculation loop to inoculate the thawed preserved bacterial strains into a shaking tube containing 5 mL of liquid LB containing corresponding antibiotics in a clean bench. 37°C, 200rpm, cultured overnight.
[0110] 3. In the ultra-clean bench, take a circle of overnight cultured bacteria and streak it onto a solid LB plate with corresponding antibiotics. Incubate overnight at 37°C.
[0111] 4. Pick a single colony on the plate for subsequent experimental operations. ...
Embodiment 3
[0162] [Example 3] Experimental results
[0163] 1. Amplification and sequence comparison of cephalosporin C acylase
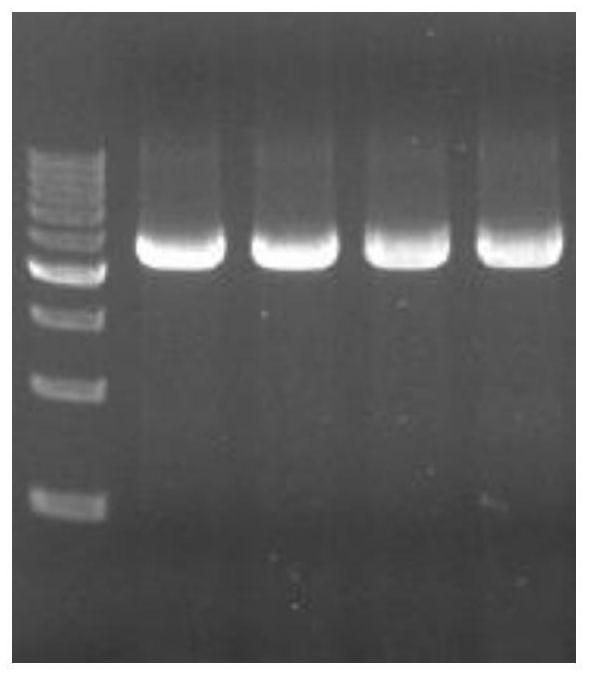
[0164] In order to obtain the cephalosporin C acylase (CA) gene, Escherichia coli BL21 (DE3) strain was activated according to the strain recovery steps described in Example 2, and the plasmid pET28α-CA was extracted. Using primers SEQ ID NO.1 and SEQ ID NO.2 (CA-1F / CA-1R) as primers to amplify the cephalosporin C acylase gene sequence, the agarose electrophoresis results of the amplified products are shown in the appendix figure 1 . The size of the PCR amplification product CA is about 2500bp, which is in line with the expected result. After the PCR product of the cephalosporin C acylase gene that met the expected results was sent to Aiji Sequencing Company for testing, the sequencing results were consistent with the target sequence results.
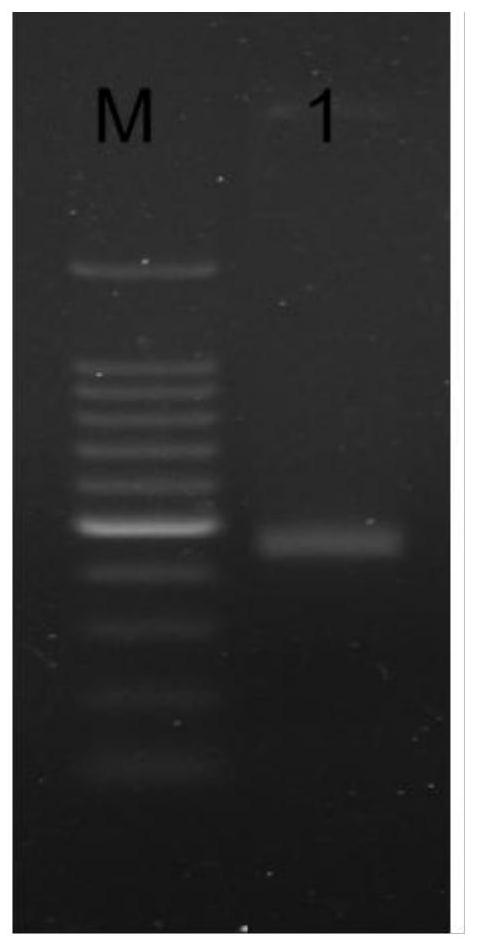
[0165] 2. Amplification and sequence comparison of the hemoglobin gene of Vibrella hyalineus
[0166] In order to...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Relative molecular mass | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com