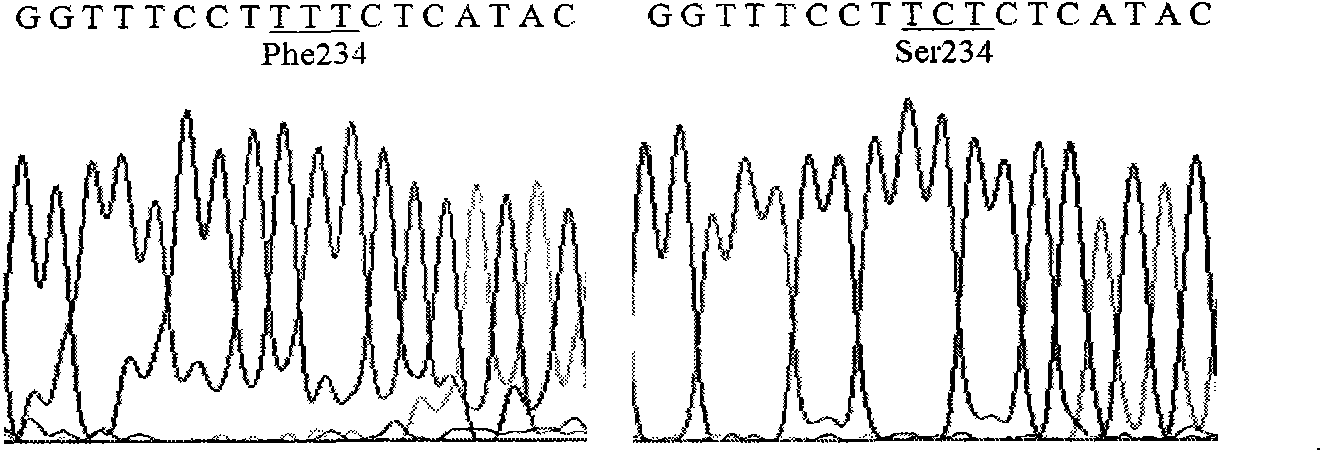
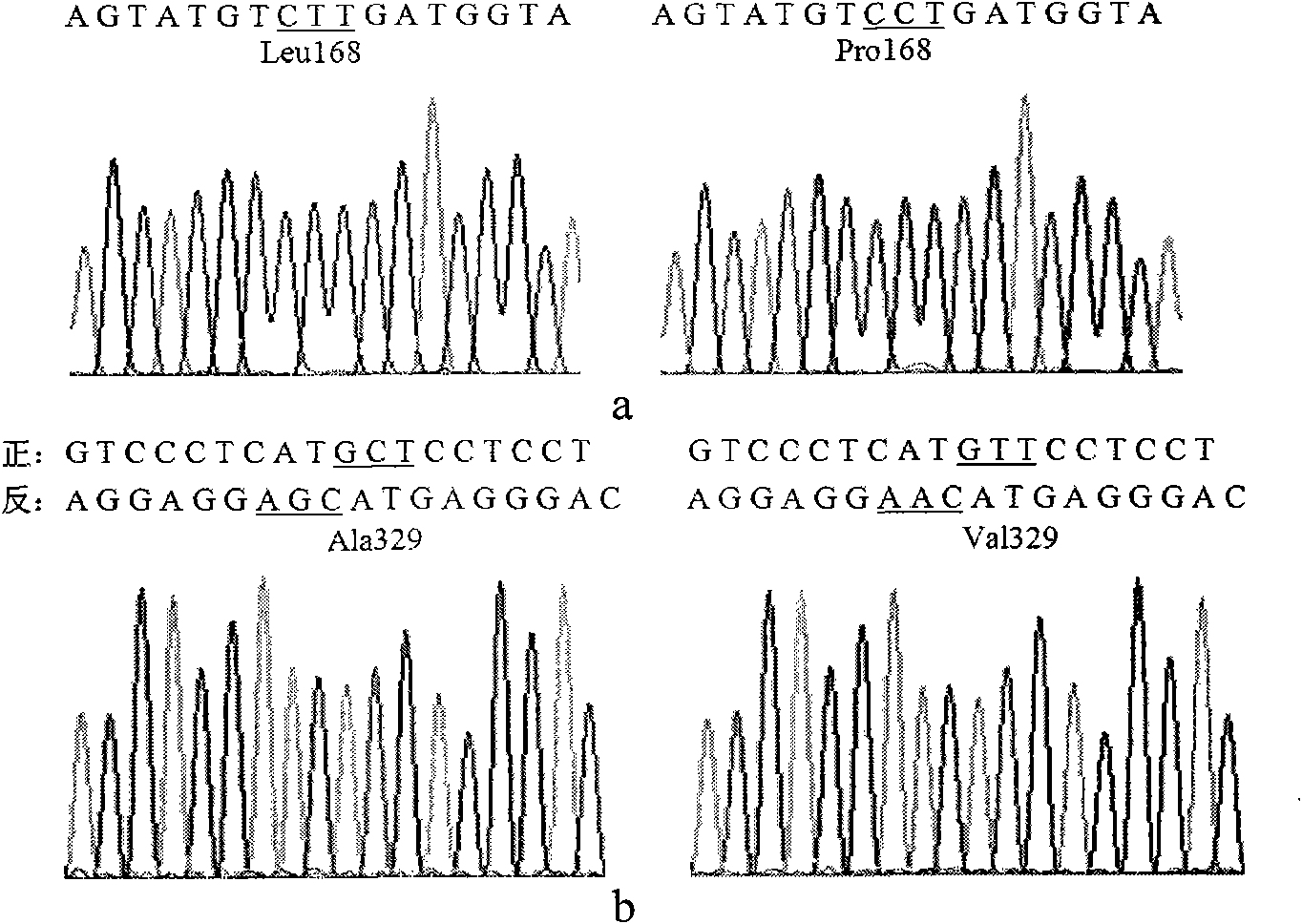
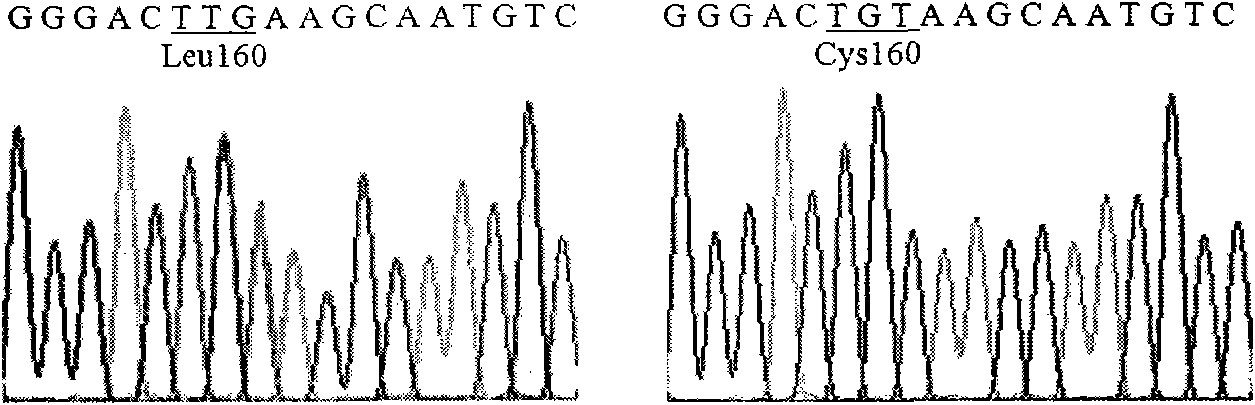
Lipase mutants with enhanced thermal stability
A thermal stability and mutant technology, applied in the field of genetic engineering of enzymes, can solve problems such as poor stability, and achieve the effect of improving thermal stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1. Construction of a Pichia pastoris library expressing lipase mutants using the error-prone PCR method
[0051] Nucleotide mutations were introduced into the lipase gene proRCL of Rhizopus sinensis in vitro by error-prone PCR. The reaction conditions for error-prone PCR are as follows:
[0052] dCTP (25mmol / L) 2μL
[0053] dTTP (25mmol / L) 2μL
[0054] dGTP (10mmol / L) 1μL
[0055] dATP (10mmol / L) 1μL
[0056] F (20pmol / μL) 1μL
[0057] R (20pmol / μL) 1μL
[0058] Mg 2+ (25mM) 14μL
[0059] mn 2+ (5mM) 1.5μL
[0060] pPIC9K-proRCL
[0061] (Plasmid carrying the Rhizopus sinensis lipase gene proRCL) 0.5 μL
[0062] Taq DNA polymerase (2.5U) 1 μL
[0063] 10×PCR buffer (without MgCl 2 ) 5μL
[0064] wxya 2 O 20 μL
[0065] Wherein, the sequences of the upstream primer F and the downstream primer R are:
[0066] F: 5'-TCAAGATCCCTAGGGTTCCTGTTGGTCATAAAGGTTC-3';
[0067] R: 5'-AATTCCAGTGCGGCCGCTTACAAACAGCTTCCTTCG-3'.
[0068] PCR amplification conditi...
Embodiment 2
[0071] Example 2, Construction of a Pichia pastoris library expressing lipase mutants using the DNA Shuffling method
[0072] The mutation sites in the lipase mutants constructed by error-prone PCR were randomly combined by DNA shuffling method. The conditions for DNA shuffling are as follows:
[0073] The genome of the Pichia pastoris library expressing lipase mutants constructed by the error-prone PCR method was extracted, digested with DNase I for 30 min, and the digested product was extracted with phenol-chloroform to remove protein and dissolved in 30 μL sterile water. Using the genome as a template, proceed as follows:
[0074] Step 1: PCR reaction system:
[0075] Taq (2.5U) 0.5μL
[0076] 5×buffer(Mg 2+ plus) 10μL
[0077] Genome 0.5 μL
[0078] dNTP (25mmol / L) 4μL
[0079] dd H 2 O 34.5 μL
[0080] PCR amplification conditions: 94°C for 3min; 94°C for 1min, 59°C for 1min, 72°C for 2min, 10 cycles; 72°C for 10min.
[0081] Step 2: Add 1 μL each of primers F a...
Embodiment 3
[0083] Embodiment 3, the screening of lipase mutant with high enzymatic activity
[0084] Copy the Pichia library stored on the MD plate to the MM plate, culture at 30°C for 2 days, and use the Pichia expressing the lipase of the parent Rhizopus sinensis as the control bacteria.
[0085] Plate initial screening: Add 200 μL of methanol to the cover of the MM plate every 12 hours to induce the expression of recombinant lipase. After 2-3 days of induction, heat the plate at 65°C for 60 minutes, cool to room temperature, and pour Fast-blue RR staining evenly on the plate. Inject about 15mL, and within 2 minutes, the dark brown colony of the monoclonal is darker than the control colony, which is the strain of primary screening purpose.
[0086] 96-well plate screening method: add 300 μL of BMGY medium to a 1.8 mL / well (flat bottom) 96-well plate, and sterilize at 121° C. for 20 minutes. Insert the strain obtained from the primary screening into it, and use the Pichia pastoris expr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com