Improved Rhizomucor miehei lipase gene and use thereof in yeast display
A technology of lipase gene and Rhizomucor miehei, applied in the field of yeast, can solve the problems of low yield and restrict the development of Rhizomucor mihei lipase enzyme preparation, etc., and achieve the effect of high catalytic performance and strong operation stability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Embodiment 1: the synthesis of improved Rhizomucor miehei lipase gene
[0029] The existing Rhizomucor miehei lipase gene is shown in SEQ.ID.NO4. The source of the gene is Rhizomucor miehei, which is different from yeast. This may be the main reason why the expression level of this gene in Pichia pastoris is not high. reason.
[0030] The present invention is based on the amino acid sequence (SEQ.ID.NO1) of Rhizomucor miehei lipase (RML), adopts the preferred codon of Pichia pastoris to replace the codon with low frequency of use of RML in yeast, and designs The gene sequence with high frequency of use in Pichia pastoris was obtained to form the following specific gene sequence, such as SEQ.ID.NO2, so as to increase the expression level of Rhizomucor miehei lipase in yeast.
[0031] The gene designed in the present invention can be artificially synthesized by PCR, and the steps are as follows:
[0032] Firstly, 46 primers are synthesized, and the primers are shown as ...
Embodiment 2
[0034] Construction of PMD18-T-RML plasmid
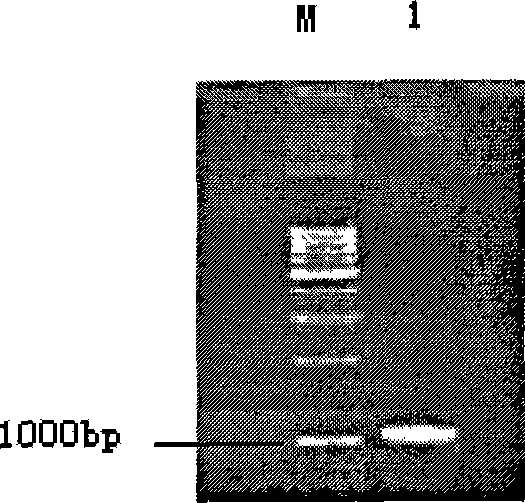
[0035] The full-length PCR product obtained in the previous step was subjected to 0.8% agarose gel electrophoresis, and the target band with a size of about 1092 bp was cut out, and the target product was purified according to the instructions of QIAGEN’s PCR gel recovery kit. Take 25.5 μL of the recovered product, and add 25 mM 1 μL of dATP, 3 μL of 10XPCR buffer, 0.5 μL of Taq enzyme, and incubated at 72°C for 30 minutes.
[0036] The product after adding A (A is adenine) was cloned by TA using the pMD18-T simple vector kit and operated according to the instructions. The 10 μL volume reaction system is as follows: T vector 1 μL (50ng), add A-added PCR product 3uL, ATP-containing 10×Buffer 1 μL, T4 DNA ligase 1 μL, make up to 10 μL with ddH2O. Slightly centrifuge, and connect overnight in a 16°C water bath. The ligation product was transformed into E.coli DH5α, and then spread on the indicator plate containing 0.5mM IPTG, 40μg / ml...
Embodiment 3
[0038] Construction of recombinant plasmid pKFS-RML
[0039] The plasmid PMD18-T-RML was used as template and RMLp1 and RMLp2 were used as primers for PCR amplification. The system is 1 μL of template; 5 μL of 10×Taq DNA polymerase buffer (containing Mg 2+ ); 2.5mmol / L dNTP 4μL; 20μM mol / L upstream and downstream primers 1μL each; Taq DNA polymerase 0.75μL, add sterile water to a total volume of 50μL. The reaction conditions are: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 45 s, annealing at 45°C for 45 s, and extension at 72°C for 2 min, a total of 30 cycles; the 30th cycle was extended at 72°C for 10 min, and the PCR product was detected by 0.8% agarose gel electrophoresis And cut the gel to recover and purify.
[0040] Both the PCR product and the pKFS plasmid were digested with EcoRI and AvrII, and the recombinant plasmid pKFS-RML was constructed with CaCl 2 Transformation method into E.coli Top10F, in Amp + Plate on LB (50mg / mL) plate and culture ove...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Vitality | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com