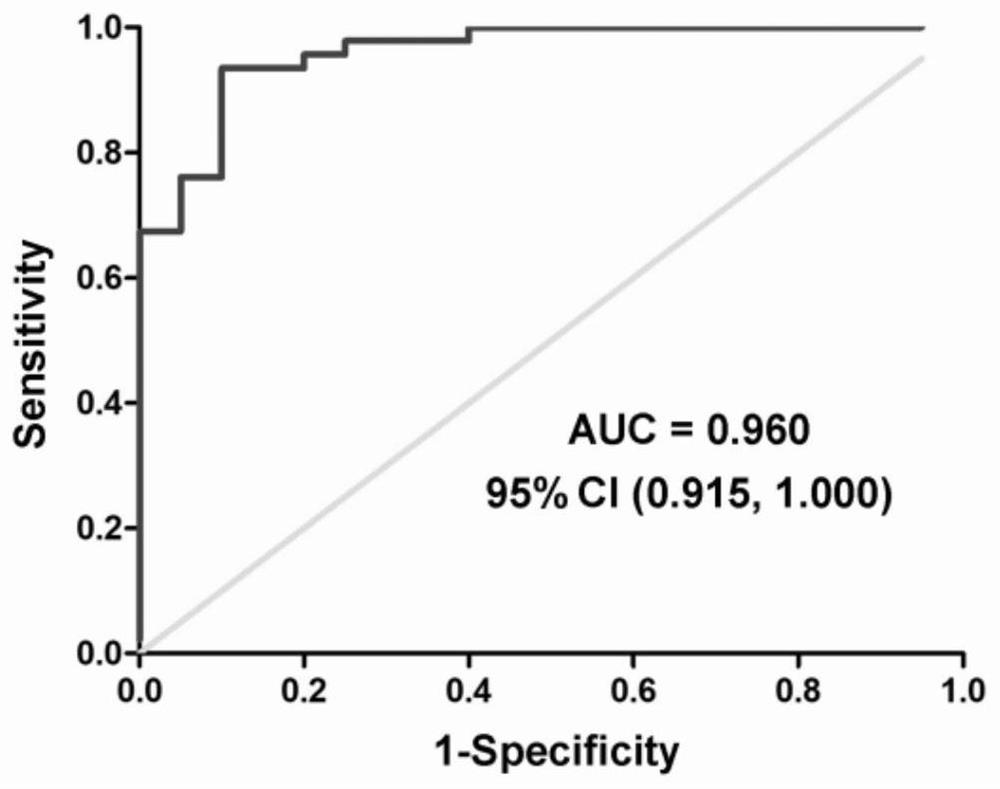
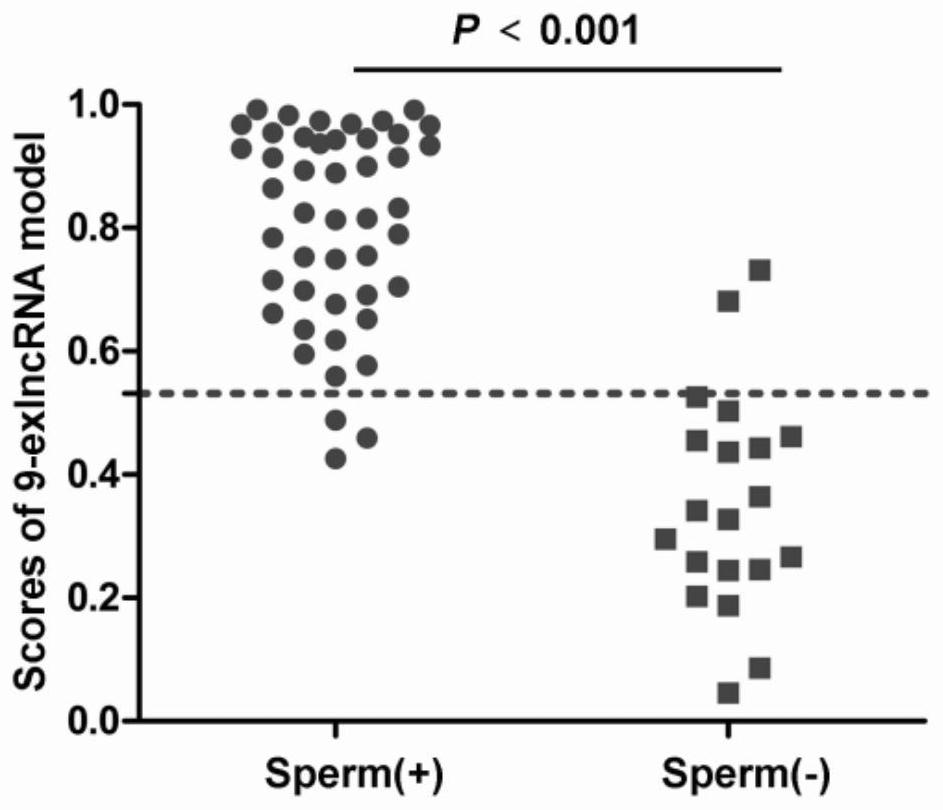
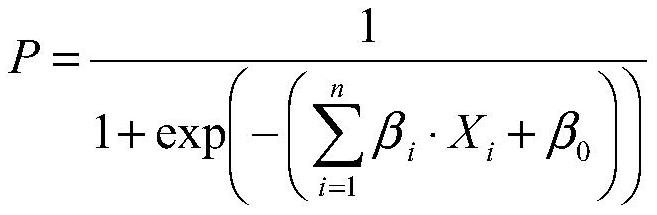A kit for predicting sperm retrieval outcome in patients with non-obstructive azoospermia
A technology for azoospermia and patients, applied in the field of biomedicine, can solve the problems of poor stability, poor sensitivity, and difficult clinical application of free RNA detection, and achieve fully scientific and practical effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1 Seminal plasma exosome purification and long-chain RNA sequencing of normal volunteers and NOA patients
[0026] Semen (>0.5mL) was taken from a single sample, centrifuged at 4°C, 12,000×g for 30min to remove cell debris, and the supernatant was collected, filtered through a 0.22μm filter, and then ultracentrifuged at 4°C, 100,000×g for 70min to collect exocrine secretions. body precipitation. The pellets were resuspended in PBS and then ultracentrifuged at 100,000 × g for 70 min at 4°C to obtain purified exosome pellets.
[0027] The purified exosomes were precipitated, added to the RNA lysis solution, and the total exosome RNA was obtained by phenol-chloroform extraction and a library was established. Followed by long-chain RNA (lncRNA) sequencing.
Embodiment 2
[0028] Example 2 Objective Gene Screening
[0029] The sequencing data in Example 1 was analyzed, and testis-specific highly expressed genes were screened according to the NCBI database information, and at the same time, the top 20 genes showing a downward trend in the sequencing results and the largest log2(FC) multiples were selected.
Embodiment 3
[0030] Example 3 Determination of lncRNA expression in seminal plasma exosomes of NOA patients
[0031] The seminal plasma exosomal RNA of NOA patients was extracted and detected by real-time quantitative polymerase chain reaction (qRT-PCR). The specific steps are as follows:
[0032] (1) Precipitate the purified exosomes obtained in Example 1, add RNA lysis solution, obtain total exosome RNA by phenol-chloroform extraction, and use Nanodrop2000 to detect the RNA concentration and purity. Perform reverse transcription reaction with 500ng RNA / 10μL system to obtain reverse transcription reaction solution and detect its concentration. The reverse transcription reaction was diluted to 10 ng / μL with DEPC water, aliquoted at 20 μL / tube and stored at -80°C. For the reverse transcription reaction solution used for qRT-PCR detection, the concentration of the reverse transcription reaction solution should be adjusted to 2.5ng / μL.
[0033](2) Using β-Actin as the internal reference gen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com