Method for constructing phage display multivariate cyclic peptide library based on disulfide bond precise pairing
A phage display and disulfide bond technology, which is applied in the field of constructing phage display multi-cyclic peptide libraries based on disulfide bond precise pairing, can solve the problems of poor tolerance to sequence modification and complex oxidation folding process, and achieve high selectivity and affinity , high oral availability and strong metabolic stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
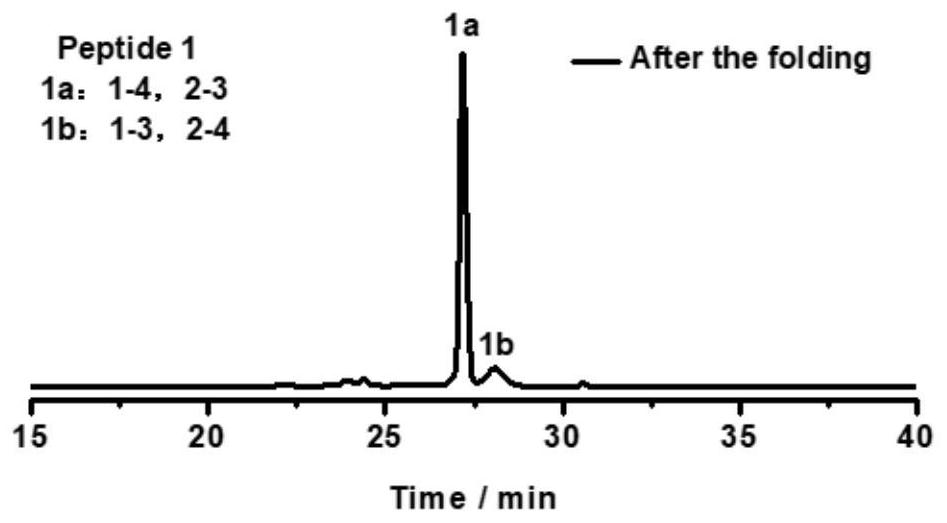
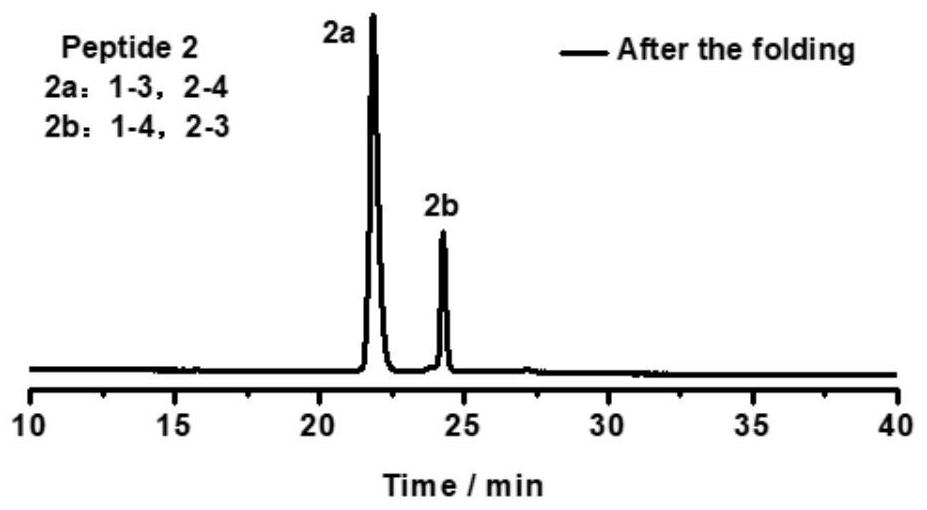
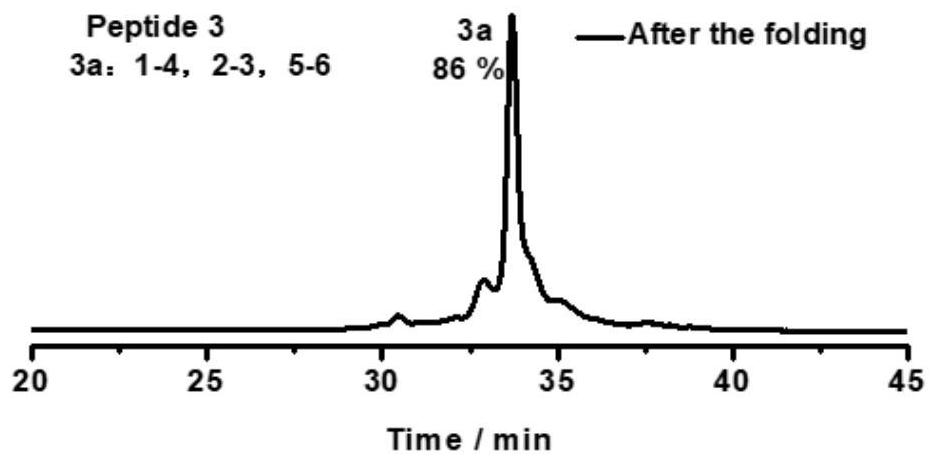
[0093] Take the oxidative folding of peptide 1 as an example (peptide 2, peptide 3, peptide 4, peptide 5, peptide 6 oxidative folding steps are the same as peptide 1, and their chromatograms are as follows Figure 2-6 ), its specific oxidation method is:
[0094] The solid powder of peptide 1 was dissolved in an aqueous solution (containing 0.1% trifluoroacetic acid to prevent oxidation of the peptide) for peptide quantification. The 50 uM polypeptide was placed in a phosphate buffer solution (100 mM pH=7.4) containing 0.5 mM GSSG, and reacted in a shaker at 37° C. for 4 h. The chromatogram of the oxidation product is as figure 1 As shown, two expected products (named 1a and 1b, respectively) were obtained.
Embodiment 2
[0096] In order to determine the pairing mode of oxidation products, taking peptide 1 as an example, a schematic diagram of the determination process is shown ( Figure 7 ). Replace the two cysteines (positions 2 and 3) in peptide 1 with Acm-protected cysteine peptide 7 (sequence: H-WGCPPC(Acm)GGKGGC(Acm)PPCGW-NH 2 ), synthesized and quantitatively placed in a phosphate buffer solution (100 mM pH=7.4) containing 0.5 mM GSSG for oxidation reaction. Utilize high-performance liquid chromatography to purify and separate the product after lyophilization, dissolve in methanol (containing 1% trifluoroacetic acid), then add twice equivalent or ten times equivalent of iodine to carry out de-Acm protection and form disulfide bond at the same time, the obtained oxidation The product is the pairing method of (1-4,2-3). The chromatographic position of the product is determined by high performance liquid chromatography, and the chromatographic position is compared with the oxidation prod...
Embodiment 3
[0098] The schematic diagram of the construction of phage display three-membered cyclic peptide library is as follows: Figure 8 shown. First, the chemically synthesized library DNA fragments and extension primers were reacted in a metal bath at 95°C for 3 minutes, and then quickly cooled on ice for 5 minutes. Afterwards, Klenow enzyme was added to the above-mentioned return system to extend the reaction for 4 hours in a 37°C metal bath, and then placed in a 65°C metal bath to inactivate the enzyme activity. Then the extension product and phagemid vector pCantab 5E were digested with SfiI (12h, 50°C) and NotI (12h, 37°C). The extension products and phagemid vectors after SfiI / NotI double digestion were purified by polyacrylamide gel electrophoresis (PAGE) and agarose gel electrophoresis, respectively. After purification, the extension products and phagemid vectors were purified at a ratio of 1:10 Molar Ratio The ligation reaction was carried out overnight (16h) at a temperat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com