SNP marker related to pepper tomato spotted wilt virus disease resistance gene as well as specific primers and application of SNP marker
A disease-resistant and specific technology, applied in the field of agricultural biology, can solve the problems of high-throughput and large-sample detection, high DNA quality requirements, unstable reaction conditions, etc., and achieve sample quality and reaction conditions. Harshness, shorten identification time, improve breeding efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Acquisition of Tsw-related SNP loci of pepper TSWV disease resistance gene and development of high-throughput KASP marker
[0041] 1. Test material
[0042] Susceptible materials: 0516 (CK-negative), eggplant door (202011C29);
[0043] Disease-resistant materials: PI152225 (CK-positive), 0516tsw (202011C35).
[0044] 2. Genomic DNA Extraction
[0045] Genome DNA was extracted from young leaves of pepper by CTAB method. For DNA extraction methods see Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA, Nucleic Acids Res 8:4321-4325.
[0046] 3. Acquisition of Tsw-related SNP loci of pepper TSWV disease resistance gene
[0047] Moury et al. (Moury B, Pflieger S, Blattes A, Lefebvre V, Palloix A. 2000. A CAPSmarker to assist selection of Tomato spotted wilt virus (TSWV) resistance inpepper. Genome 43, 137–142.) developed a pair of resistance to Tsw Genetically tightly linked SCAR / CAPS marker SCAC 568 , but only the traditional C...
Embodiment 2
[0063] Example 2 Method for detecting pepper TSWV resistance gene Tsw using high-throughput KASP marker Pep(Tsw)-k497
[0064] 1. PCR amplification
[0065] The young leaves of pepper to be tested were taken as samples, and the genomic DNA was extracted from the samples by CTAB method. The above primers 1, 2 and 3 were used to pair susceptible material 0516, eggplant and disease-resistant materials PI152225, 0516tsw and their two F 1 Substitute samples for PCR amplification, and the reaction system is carried out in a 96-well plate.
[0066] PCR reaction system for KASP genotyping used for amplification (10 μl system):
[0067] Table 1 Reaction system
[0068] Element Dosage KASP V4.0 2×Master Mix 5μl KASP 72×assay mix 0.14μl genomic DNA 10ng wxya 2 o
Add to the total amount of the system to 10ul
[0069] Among them, KASP V4.0 2×Master Mix consists of fluorescent probe A, fluorescent probe B, quenching probe A and quenching p...
Embodiment 3
[0084] Example 3 Application of high-throughput KASP marker in backcross breeding of pepper TSWV disease resistance gene Tsw
[0085] 1. Materials to be tested:
[0086] 6 pepper hybrid populations including 3 BCs 3 F 2 Population (3 generations of backcrossing, 2 generations of selfing), 3 BCs 4 colonies, each colony contains 14 to 16 individual plants (see Table 2 for details). Population construction was based on three pepper inbred lines (respectively, Lujiao M, Zhu Dachang, 0818) as recurrent parents after crossing with TSWV disease-resistant material 0516Tsw, using the recurrent parents to backcross for 3-4 generations, and then selfing 0-2 generations. obtained on behalf of.
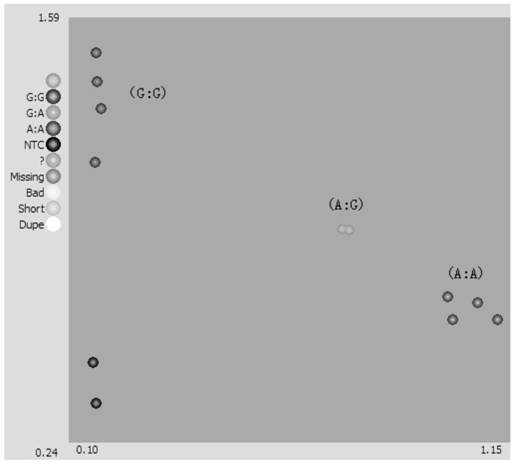
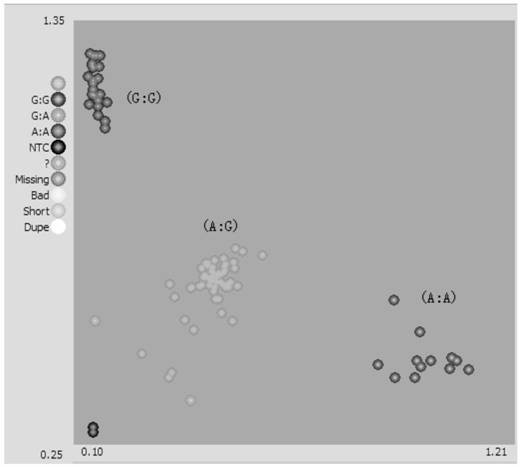
[0087] With reference to the method in Example 2, the high-throughput KASP marker was used to identify the genotype of the pepper Tswv resistance gene Tsw for individual plants in the population (the results are shown in Table 2 and Table 2). image 3 ).
[0088] Table 2 Genotype detection r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com