Double-fluorescent-protein positioning detection system for detecting mitochondrial autophagy of cells and application
A technology of mitophagy and dual fluorescent protein, applied in the biological field, can solve the problems of cumbersome process, cumbersome electron microscope detection process, long cycle, etc., and achieve the effect of solving the cumbersome operation process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
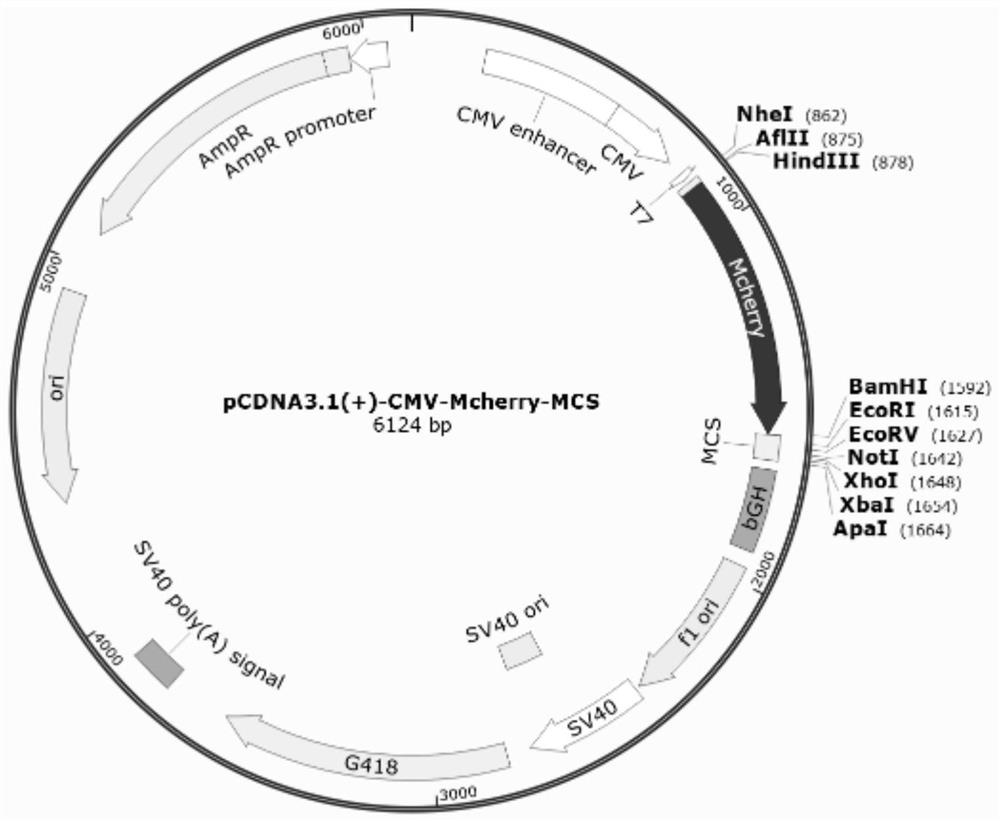
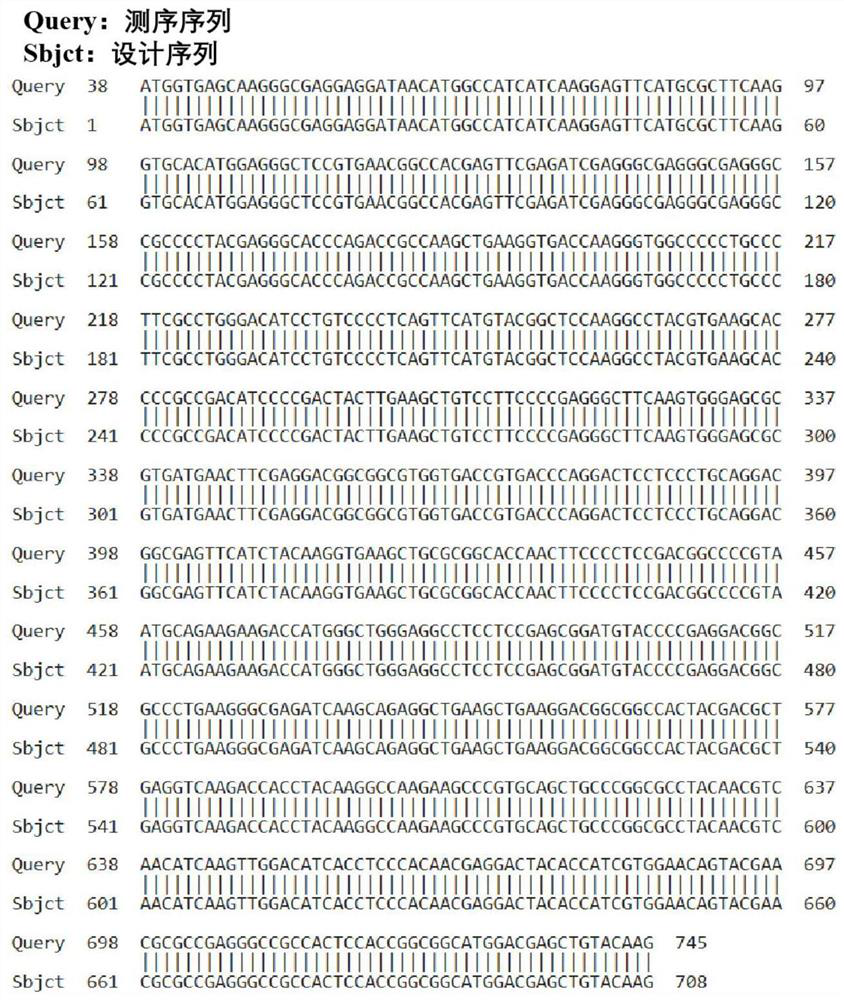
[0077] The pcDNA3.1(+) empty plasmid was used to design Mcherry primers, the PCR products were tapped and recovered, and the pcDNA3.1(+)-Mcherry-MCS fusion expression plasmid was constructed, identified by enzyme digestion, sequence comparison, and expression identification.
[0078] Using the sequence of the PcDNA3.1-Tom20-Mcherry plasmid as a template (remove the TAA stop codon), select the HindⅢ and BamhI restriction sites on the pcDNA3.1(+) empty plasmid, and use the Primer 5 software to design Mcherry primers , and add the corresponding restriction site sequence (HindⅢ: AAGCTT , Bamh I: GGATCC ) and the corresponding protected base sequence (HindⅢ: CCC, BamhⅠ: CG), as follows:
[0079] Mcherry-HindⅢ-F: CCC AAGCTT ATGGTGAGCAAGGGCGAGG; (SEQ ID NO: 3)
[0080] Mcherry-BamhⅠ-R: CG GGATCC CTTGTACAGCTCGTCCATGCC; (SEQ ID NO: 4)
[0081]The target fragment (725 bp) was recovered by 1% agarose gel electrophoresis after PCR reaction of 50 μL system, and the target fragment...
Embodiment 2
[0086] Using the pcDNA3.1(+)-Mcherry-MCS fusion expression plasmid and designing the full-length primer of LC3B, using the cDNA of human lung adenocarcinoma cell line A549 as a template, the PCR product was recovered by tapping rubber to construct pcDNA3.1(+)-Mcherry- Lc3b fusion expression plasmid, enzyme digestion identification, sequencing comparison verification, expression identification.
[0087] Referring to the sequence information of the CDS coding region of Homo sapiens microtubule associated protein 1light chain 3beta (NCBI Reference Sequence: NM_022818.5), using the cDNA of human lung adenocarcinoma cell A549 as a template, select pcDNA3.1(+)-Mcherry-MCS fusion expression EcorI and XbaI restriction sites on the plasmid, use Primer 5 software to design MAP1LC3B primers, and ensure that LC3B will not undergo frameshift mutations (base T is added to the 5' end of the forward primer), and in the forward and reverse Add the corresponding restriction site sequence (EcorI...
Embodiment 3
[0095] Using PcNA3.1-MCS-EGFP fusion to express the empty plasmid, design the full-length primer of PHB1, use the cDNA of human lung adenocarcinoma cell line A549 as a template, and recover the PCR product by rubber tapping to construct the fusion of pcDNA3.1(+)-PHB1-EGFP Expression plasmids, enzyme digestion identification, sequencing comparison, expression identification.
[0096]Referring to Homo sapiens prohibitin (PHB), transcript variant 2 (NCBI ReferenceSequence: NM_002634.4) CDS coding region sequence information (remove stop codon TGA), using the cDNA of human lung adenocarcinoma cell A549 as a template, select pcDNA3.1 ( +)-MCS-EGFP fusion expresses the HindⅢ and EcorⅠ restriction sites on the empty plasmid, and uses Primer 5 software to design MAP1LC3B primers, and ensures that EGFP will not undergo frameshift mutations (add Base C), and add the corresponding restriction site sequence (HindⅢ: AAGCTT, EcorI: GAATTC) and the corresponding protection base sequence (Hin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com