A pathogenic strain causing patchouli bacterial wilt and its application
A technology of patchouli and bacterial wilt, which is applied to the field of pathogenic strains that cause patchouli bacterial wilt, and achieves a high affinity effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Isolation and purification of Patchouli solanacearum and pathogenicity assay
[0037] (1) Separation and purification: collect patchouli plants with typical symptoms of bacterial wilt disease from the field, soak the diseased plants in tissue, inoculate the infusion solution on a TTC plate, culture at 30°C for 48 hours, and pick out bacteria that are related to bacterial wilt Single colonies with similar colony morphology were purified and preserved.
[0038]Among them, the culture medium of TTC plate is acid hydrolyzed casein 1.0g / L, glucose 5.0g / L, peptone 10.0g / L, agar powder 10g / L and TTC 0.05g / L, pH7.2.
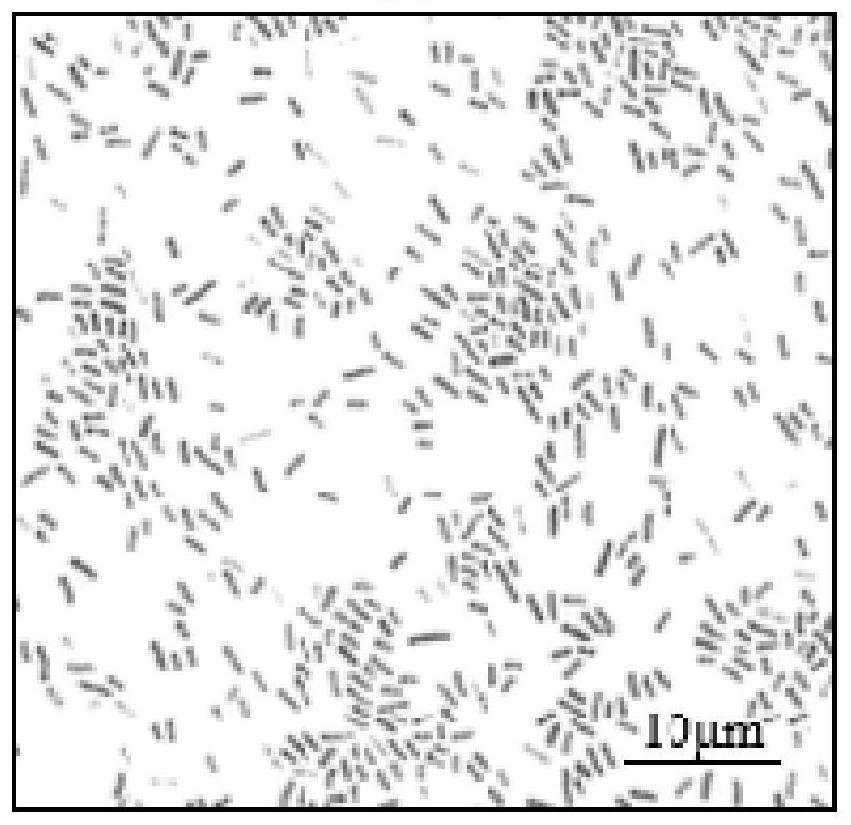
[0039] (2) Observation of colony morphology: inoculate the purified and preserved strains on a TTC plate by the three-line method, culture at 30° C. for 24 hours, and observe the bacterial colony morphology. The colonies are smooth circular bulges, with a red center and a white bacterial circle, with obvious fluidity, such as figure 1 shown.
[0040] ...
Embodiment 216
[0041] Example 2 16s rDNA sequence identification
[0042] Genomic DNA of strain Pa82 was used as a template to perform PCR amplification of 16s rDNA gene. The primers were: 27F: 5′-AGAGTTTGATCCTGGCTCAG-3′, 1541R: 5′-AAGGAGGTGATCCAGCCGCA-3′. Use 25 μL system for PCR reaction: 1 μL each of 27F / 1541R (10 μM), 1.5 μL of genomic DNA, ddH 2 O 9.0 μL, Taq enzyme 12.5 μL; reaction parameters: pre-denaturation at 94°C for 2 min; denaturation at 94°C for 30 s, annealing at 58°C for 30 s, extension at 72°C for 100 s, 35 cycles; extension at 72°C for 10 min. PCR samples were subjected to agarose gel electrophoresis ( Figure 4 A), and sequence the samples. Compare the sequence results of the 16s rDNA gene on NCBI, select strains with high similarity and other plant pathogenic bacterial strains, and use the Neighbor-joining method in MEGA5.1 software to construct the phylogenetic tree of 16s rDNA, as shown in Figure 5 shown. The results showed that the strain Pa82 was clustered with ...
Embodiment 3
[0044] Example 3 Identification of rpoB gene sequence
[0045] Genomic DNA of strain Pa82 was used as a template to perform PCR amplification of rpoB gene, and the primers were: rpoB F: 5′-ATGGTTTACTCCCTATACCGAGA-3′, rpoB R: 5′-TTACTCGTCTTTCCAGCTCGAT-3′. PCR reaction uses 25 μL system: rpoB F / R primer (10 μM) 1 μL each, genomic DNA 1.5 μL, ddH 2 O 9.0 μL, Taq enzyme 12.5 μL; reaction parameters: pre-denaturation at 94°C for 2 min; denaturation at 94°C for 30 s, annealing at 58°C for 30 s, extension at 72°C for 100 s, 35 cycles; extension at 72°C for 10 min. PCR samples were subjected to agarose gel electrophoresis ( Figure 4 B), and sequence the samples. Compare the sequence results of the rpoB gene on NCBI, select strains with high similarity and other plant pathogenic bacterial strains, and use the Neighbor-joining method in the MEGA5.1 software to construct a phylogenetic tree of the rpoB gene, as shown in Figure 6 shown. The results showed that the strain Pa82 was cl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com