Application and method of bombyx mori amylase gene BmAmy1
An amylase, silkworm technology, applied in genetic engineering, biochemical equipment and methods, enzymes, etc., can solve problems such as functions to be studied, and achieve the effect of improving the weight of the whole cocoon and important production and application potential
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1, Identification, cloning and expression profile analysis of the silkworm BmAmy1 gene
[0033] 1. Identification of silkworm α-amylase gene (BmAmy1)
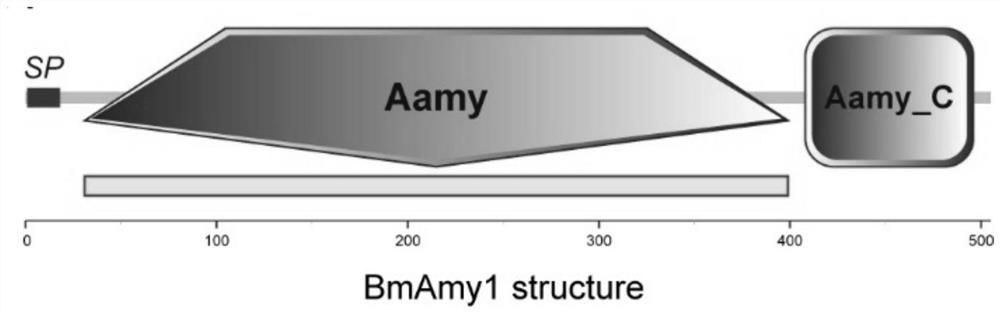
[0034] Download the reported amino acid sequences of silkworm α-amylase genes and other species α-amylase genes from NCBI as search seed sequences, and use the silkworm genome database (SilkDB) to carry out Blast comparison and identification (Blast parameter is set to Blastp) to obtain candidates sequence. Then the candidate sequences were manually screened, and finally the domains were predicted on the SMART website (http: / / smart.embl-heidelberg.de / ) to obtain the nucleic acid and protein sequences of BmAmy1.
[0035] 2. Multiple sequence alignment and phylogenetic analysis of BmAmy1
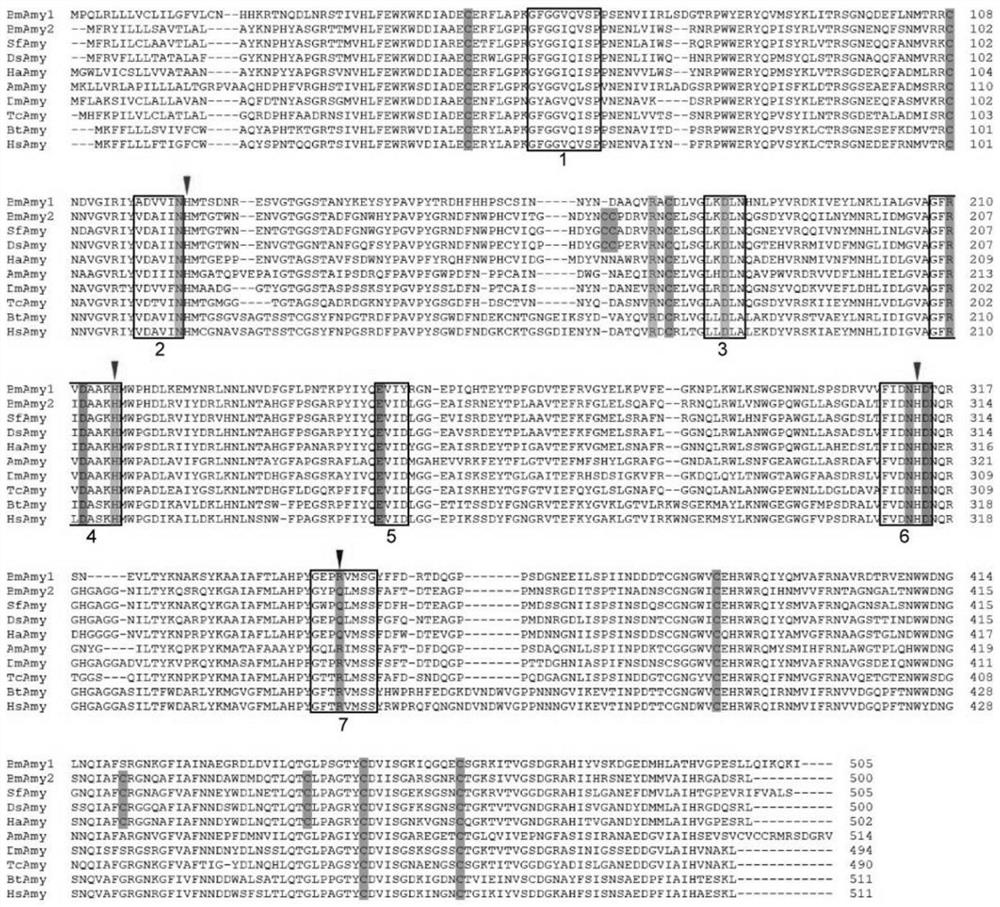
[0036] According to the reported literature and NCBI database search, the protein sequences of α-amylase genes of multiple species were downloaded, multiple sequence alignments were performed using Clustalx, and the files were ...
Embodiment 2
[0044] Embodiment 2, the expression characteristic analysis of BmAmy1 gene
[0045] The expression characteristics analysis of BmAmy1 was carried out by fluorescent quantitative PCR method, and the fluorescent quantitative PCR primers of BmAmy1 were BmAmy1-qRT-F: 5'-ccatcatccgtcctgctctat-3' (SEQ ID NO.4) and BmAmy1-qRT-R: 5'-ggcaagttgtgattcaagtcct -3' (SEQ ID NO. 5). The internal reference gene fluorescent quantitative PCR primers were sw22934-F: 5'-ttcgtactggctcttctcgt-3' (SEQ ID NO.6) and sw22934-R: 5'-caaagttgatagcaattccct-3' (SEQ ID NO.7). The qPCR experiment used the SYBR Premix Ex Taq II kit from Takara, and the fluorescent quantitative PCR instrument was qTOWER in Jena, Germany 3 touch quantitative PCR instrument. The PCR program was as follows: pre-denaturation at 95°C for 30s, followed by denaturation at 95°C for 3s, annealing at 60°C for 30s, and 40 cycles. Three experiments were carried out for each sample, and the Ct value was collected for data analysis. thro...
Embodiment 3
[0046]Example 3, Eukaryotic expression, purification and activity detection of the silkworm BmAmy1 gene
[0047] For eukaryotic expression, Pichia pastoris (Pichia pastoris) X-33 strain was selected. X-33 Pichia pastoris contains the AOX1 gene, and the resulting transformant is Mut + genotype. The expression vector is pPICZαA vector containing Zeocin resistance.
[0048] (1) Yeast eukaryotic expression vector construction and transformation
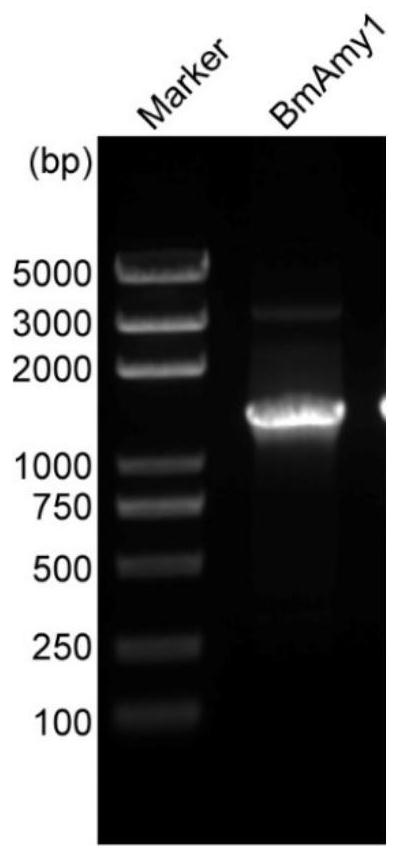
[0049] According to the sequence characteristics of BmAmy1 and the structural characteristics of the pPICZαA vector, a gene-specific primer EcoRI-BmAmy1-F with EcoR I and Not I restriction sites was designed: 5’-cg gaattc aatcatcataaaaggacgaacc-3' (SEQ ID NO.8) and NotI-BmAmy1-R: 5'-attt gcggccgc tatcttctgcttgatctggag-3' (SEQ ID NO. 9). Using the previously constructed T5-BmAmy1 vector as a template, the gene fragment with EcoRI and Not I restriction sites was amplified by PCR with TransTaq HiFi DNA polymerase. The gel was cut and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com