Preterm birth risk prediction using change in blood microbial community
A microorganism and risk technology, applied in the field of compositions for diagnosing the risk of preterm birth, can solve problems such as non-specificity and insensitive methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077]Example 1: Selection of experimental materials
[0078]Analysis of microbiological groups in the blood of un pregnant women, pregnant pregnant women and pregnant pregnant women, selected 41 unsophiliated women in the Pear Flower Women's University Wooden Hospital, 20 pregnant women and 21 pregnant women in pregnant women. The normal foot-filled childbirth group was selected for pregnant women who experienced the feet childbirth after 37 weeks of pregnancy. When the pregnant woman occurs or the symptoms of fetal membrane, the parent blood is collected in a test tube containing EDTA, and plasma is separated and stored at -70 ° C.
[0079]Only the single-weef production data of 25-42 weeks, excluded multi-child, dead production, malformed, chronic hypertension, placenta preamp, and placental peeling.
[0080]Regardless of pregnant women from healthy women without pregnancy-related diseases.
[0081]All processes and processes of the experiment were approved by the institutional review commit...
Embodiment 2
[0082]Example 2: DNA extraction and 16S rRNA sequencing
[0083]Bacterial DNA was extracted from the blood samples of Example 1 using the PowerMax Soil DNA separation kit (Mobio, Carlsbad, CA, US) using the PowerMax Soil DNA separation kit (Mobio, Carlsbad, CA, USA).
[0084]The 16S RRNA gene V3-V4 high variable region (519F-816R) of the bacterial genomic DNA (519F-816R) was amplified by the ILLUMINA 16S Acer. The fusion primer sequence used for amplified strip codes is as follows
[0085]16S_V3_F primers (SEQ ID NO: 1)
[0086]5'-tcgcggcagcgtcagatgtgtataagagagacagcctacgggnggcwgcAg-3 '
[0087]16S_V4_F primers (SEQ ID NO: 2)
[0088]5'-gtctcgtgggctcgggagatgtgtataagagagaggactachvggtatctaatcc -3.
[0089]The library is prepared using the PCR product in accordance with the MISEQ System Guide (Illumina) and quantitatively quantitatively using Qiaxpert (Qiagen, Hilden, Germany). After extracting and quantifying the PCR product, the equimolar ratio of each sample was analyzed using Miseq (Illumina) to analyze...
Embodiment 3
[0090]Example 3: Analysis of bacteria composition
[0091]The sequence information obtained in Example 2 was treated using Miseq (Illumina) according to barcode algorithm and primer sequence. Use the analysis program MDX-Pro Ver.1 (MD Healthcare, Seoul, South Korea) to classify (TAXOMICASSIGNMENT).
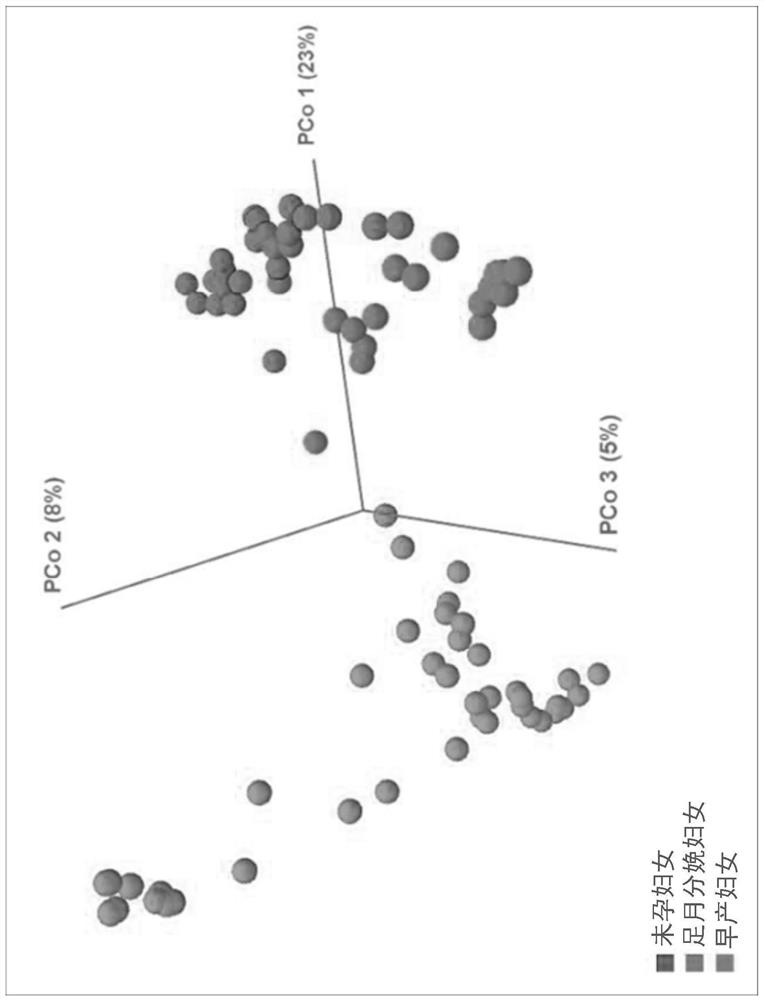
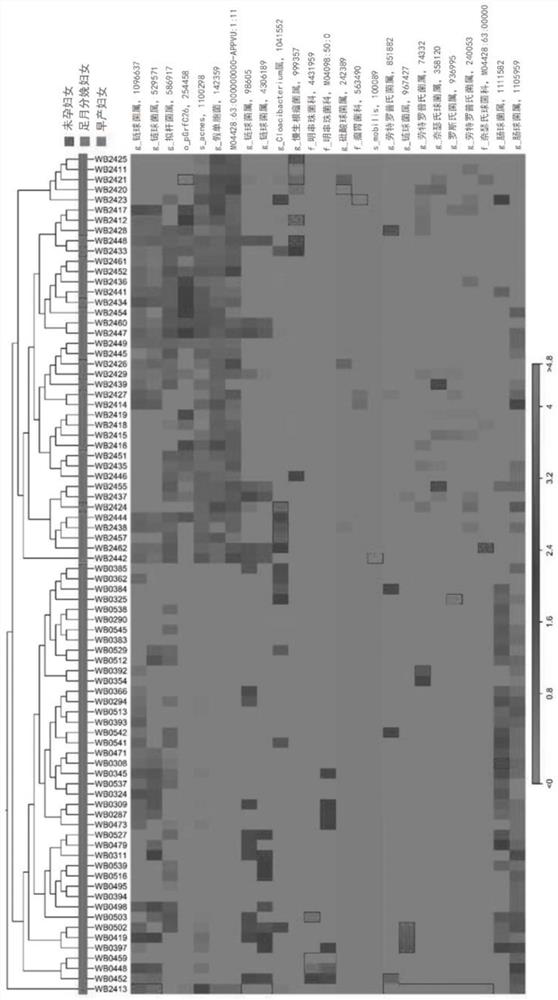
[0092]For the result of the serial number analysis, the high quality reading is selected after the reading length (= 300 bp) and the quality score (the average Phred Score = 20) are selected. Use the sequence clustering algorithm CD-HIT to cluster the operation classification unit. Subsequently, UCLST and QIIME are classified using Uclst and QIIME according to the 16SRNA sequence database of Greengenes 8.15.13. All 16SRNA sequences are allocated to the classification level based on the sequence similarity.Figure 4a to Figure 4fBy stacking the strip of the problem of 6 bacterial communities in the blood of women, women in PTB women and PTB women. If a sequence or redundant sequence is lacking ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com