Method for efficiently synthesizing mequinomycin A in streptomyces albus
A technology of Streptomyces albicans and moronomycin, which is applied in the directions of microorganism-based methods, botanical equipment and methods, biochemical equipment and methods, etc. Create job problems, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0058] Construction of Streptomyces albus LX03 High-Producing Strain of Moenomyces A
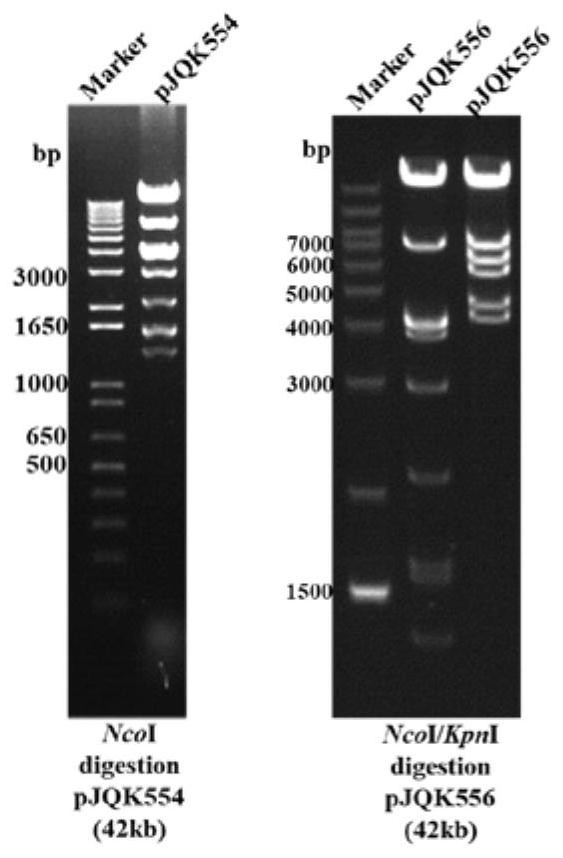
[0059] Step 1: Construction of the fosmid plasmid pJQK455 containing the moenomycin biosynthesis gene cluster cluster 1; through antiSMASH online analysis, an incomplete moenomycin biosynthesis gene was found in the whole genome of Streptomyces lincoeum (GenBank: CP016438.1) Cluster cluster 1 (GenBank assembly accession: GCA_003344445.1RefSeq assembly accession: GCF_003344445.1). The designed primers (Moe-L-F / Moe-L-R and Moe-R-F / Moe-R-R) were screened by polymerase chain reaction shrinkage genome library method to obtain fosmid plasmids 5B1, 8F3, 9A8 containing moenomycin biosynthesis gene cluster and 16G7 were confirmed to be correct by restriction enzyme digestion with BamHI and KpnⅠ, and 5B1 was selected for sequencing and confirmed to be the correct plasmid, named pJQK455 ( Figure 5 ).
[0060] The primers used in the above step 1 are:
[0061] Primer name base sequence ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com