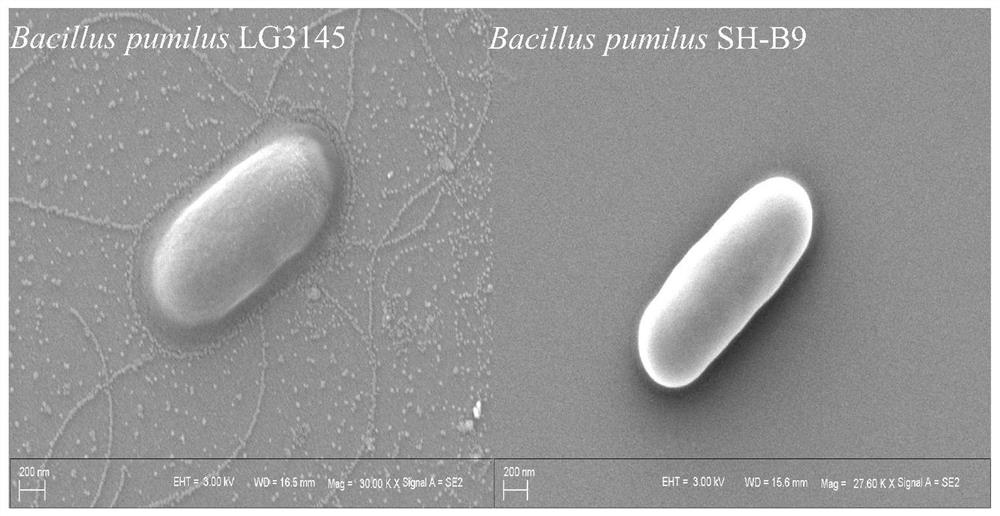
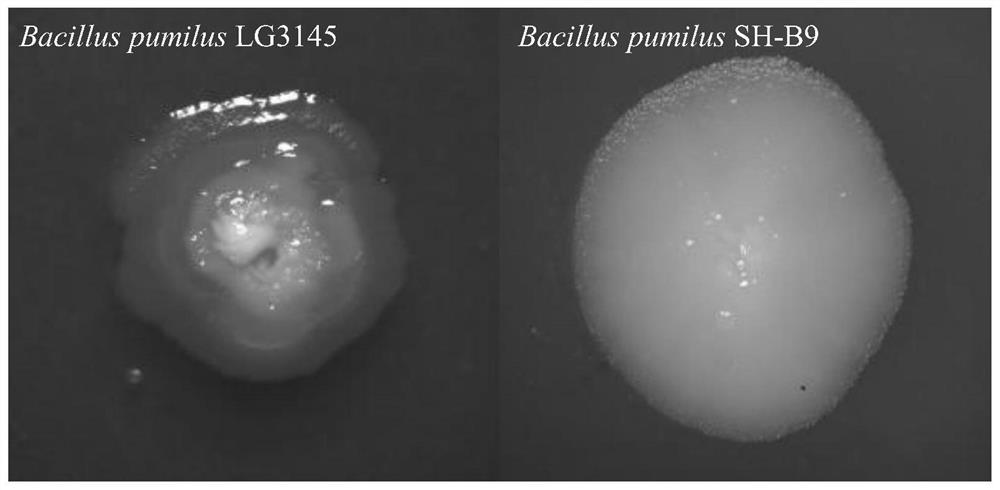
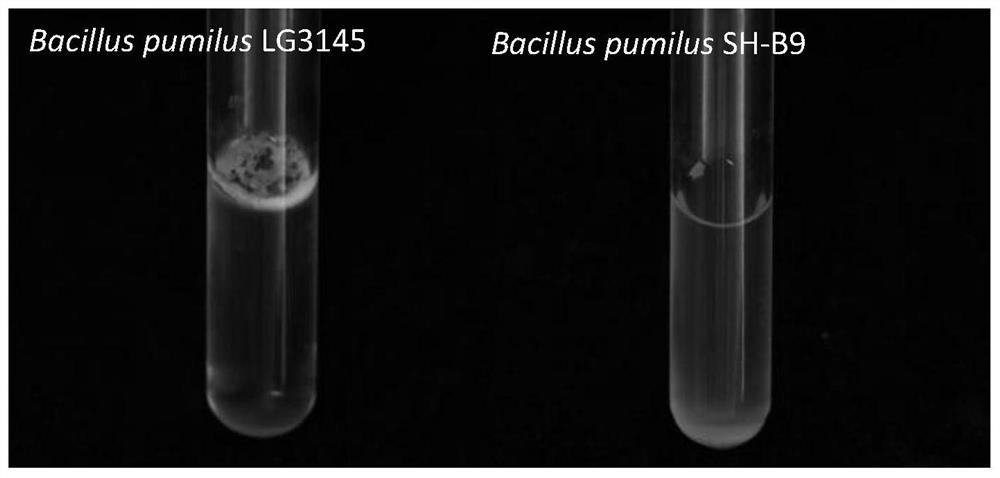
Construction method of glucose-tolerant high-secretion genetic engineering recipient bacterium
A technology of genetic engineering and construction method, applied in the field of genetic modification and construction of glucose-tolerant and high-secretion genetically engineered receptor bacteria, can solve the problems of genetic and unstable endotoxin contained in the engineered receptor bacteria, and achieve large yield and increased secretion. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] 1. Construction of recombinant plasmids
[0024] 1. Design of gRNA
[0025] Determine the target gene that has a CCR effect and contains the cis element of crebox in the sugar metabolism pathway, specifically select the following seven target genes:
[0026] ackA UP12_RS13195 acetate kinase
[0027] ycgE UP12_RS01730 aminotransferase
[0028] ptsH UP12_RS06700 phosphocarrierprotein HPr
[0029] budA UP12_RS16915 acetolactate decarboxylase
[0030] acsA UP12_RS13395 acetyl-CoA synthetase
[0031] xylA UP12_RS09365 xylose isomerase
[0032] sucC UP12_RS07835 succinate-CoAligase[ADP-forming]subunitbeta
[0033] Determine the crebox sequence and specific location in the target gene.
[0034] After positioning with gRNA, the Cas9 protein cuts about 3bp at the 5' end of the binding site. Select the cre box and the subsequent 20bp sequence and select PAM as NNG to design the target fragment. Each gene corresponds to multiple Group possible target fragments, summarize t...
Embodiment 2
[0093] Glucose fermentation medium (GYN): 2g yeast powder, 0.05g MgSO4·7H2O, add 1% volume of urea mother solution (20% 0.05Mpa sterilization for 20min), add glucose according to different concentrations. Add deionized water to 100mL, pH 7.2-7.5, sterilize at 0.08Mpa for 20min.
[0094]Pick the recombinant Bacillus pumilus LG3145 and culture it in 4 μg / ml liquid medium containing chloramphenicol for 12 hours, then take 200 μL and add it to 100 ml glucose fermentation medium at 150 rpm, shake and culture at 37°C for 24 hours to observe the growth status. And use the ultraviolet spectrophotometer to take samples every two hours to measure the bacterial concentration. Bacillus pumilus LG3145 grows much better than wild bacteria in the case of high sugar.
[0095] When the recombinant Bacillus pumilus LG3145 was cultured in different concentrations of glucose fermentation medium, the sugar tolerance of Bacillus pumilus LG3145 was significantly improved, and the sugar tolerance cou...
Embodiment 3
[0101] Determination of Extracellular Protein Activity of Recombinant Bacillus pumilus
[0102] 1. Preparation of double-layer skimmed milk powder solid medium: lower layer: 2.5g of agar was dissolved in 100ml of deionized water, autoclaved at 121°C for 30 minutes, and the solution was spread on the bottom of the petri dish and allowed to cool and solidify. Upper layer: 2g of skimmed milk powder, 2.5g of agar dissolved in 100ml of deionized water, autoclaved at 105°C for 20 minutes to obtain 2% milk medium, spread the solution on the solidified lower layer of agar, let cool and solidify.
[0103] 2. Pick a single colony from the plate, inoculate it into a test tube containing 5mL LB medium, culture overnight at 37°C, 120-200r / min, transfer 1mL of the bacterial solution to a 250mL Erlenmeyer flask inoculated with 100mL LB medium , 37°C, 120-200r / min culture. Take 1ml every two hours, centrifuge at 12000r for 5min, get the supernatant, take 10ul supernatant and spot on the plat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com