Kit and method for detecting lumpy skin disease viruses
A technology for nodular and skin diseases, applied in nucleic acid detection kits and detection fields for bovine nodular skin diseases, can solve problems such as poor sensitivity, and achieve short amplification time, multi-data support, and high efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Embodiment 1: the preparation of positive template
[0063] The positive cloned plasmids were synthesized and sequenced by Beijing Liuhe Huada Gene Co., Ltd. After the sequencing results were compared with the LSDV-LD126 and LSDV001 gene sequences published on GenBank, the concentration and purity of the plasmids were measured by an automatic microplate reader, and named respectively. For LSDV-LD126, LSDV001.
[0064] The LSDV-LD126 gene sequence is shown in SEQ ID NO: 1, specifically:
[0065] ATGGGAATAGTATCTGTTGTATACGTCGTAGTACCATTTTCATTTATTGTTTTACTTTCATATATATTTTTTGAATACAAAAATGTTATTAAAAAAATGTTATTTAAAAAAAAAGGGAAAAATCAAAAAAGAACATGTGTTAGACTTAATTCAATCACTTATTCAACAAATAGTATAAAATCCACTATATCAGAAAGTACTTGGTCAAATTGTAGTAATGATACATTTGTAAAAAATGAAAAAGAAAATGTAGAGATTGTTGAAATTAAACGTTGTGATAATGAATTAATTGAAGAAAGTAATAATAACGTTTTAGAAAATGGATGTACCACAAATACAGGTGAAGAAAATTTAATTTGGGACGATAACAACGTTTATGATTTACCACCTAATGATAGTGTTTATGATTTACCACCTAACGATTTGAGTTGTAACAACGATTGTGTTTATACATTACCGGATGACAATGTTTCAAACATAGAGG...
Embodiment 2
[0070] Example 2: Design and synthesis of primers and probes
[0071] According to the LSDV genome sequence (AF325528.1) published on GenBank, the genome sequence was analyzed and compared using DNAMan software, and primers and probes were designed for the conserved region of the genome sequence. The sequence is shown in Table 1. Primers and probes were synthesized by Shanghai Shining Molecular Biotechnology Co., Ltd.
[0072] Table 1: Primers and Probes
[0073]
Embodiment 3
[0074] Example 3: Identification of Primer Specificity
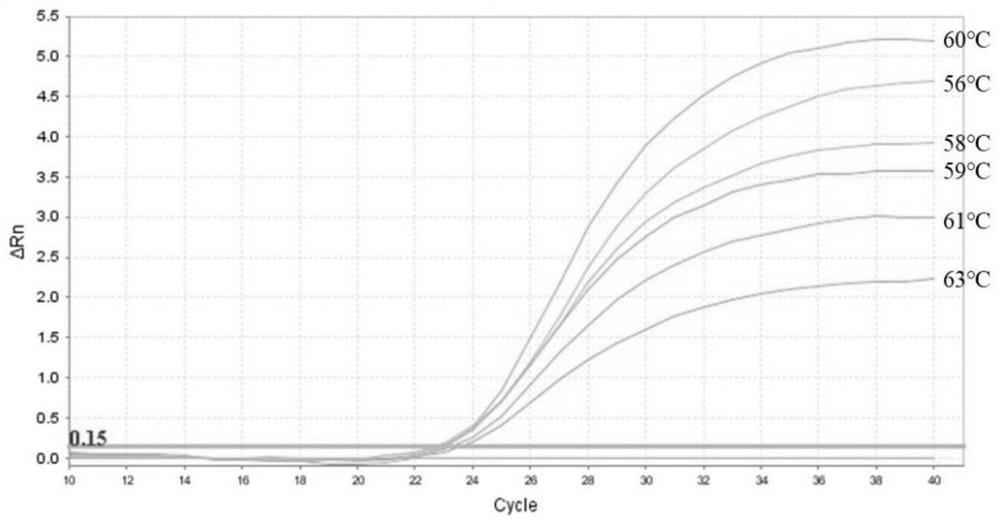
[0075] LSDV-LD126 and LSDV001 positive recombinant plasmids were prepared according to the reaction system provided by the instructions of QuantiNova SYBR Green PCR Kit (CatNo.208052), that is, 10 μL of Premix Taq (2×) mixture, 0.1 μL of QN ROX ReferenceDye, 1.4 μL of each upstream primer and downstream primer ( 0.7μM), template DNA 1μL, with ddH 2 O was supplemented to 20 μL for amplification. The amplification conditions were pre-denaturation at 95°C for 2 minutes; 40 cycles of amplification at 95°C for 5s and 60°C for 10s. After the amplification, continue to do the melting curve, 95°C, 15s, 60°C, 1min; 95°C, 30s, 60°C, 15s. Verify the specificity of the primers.
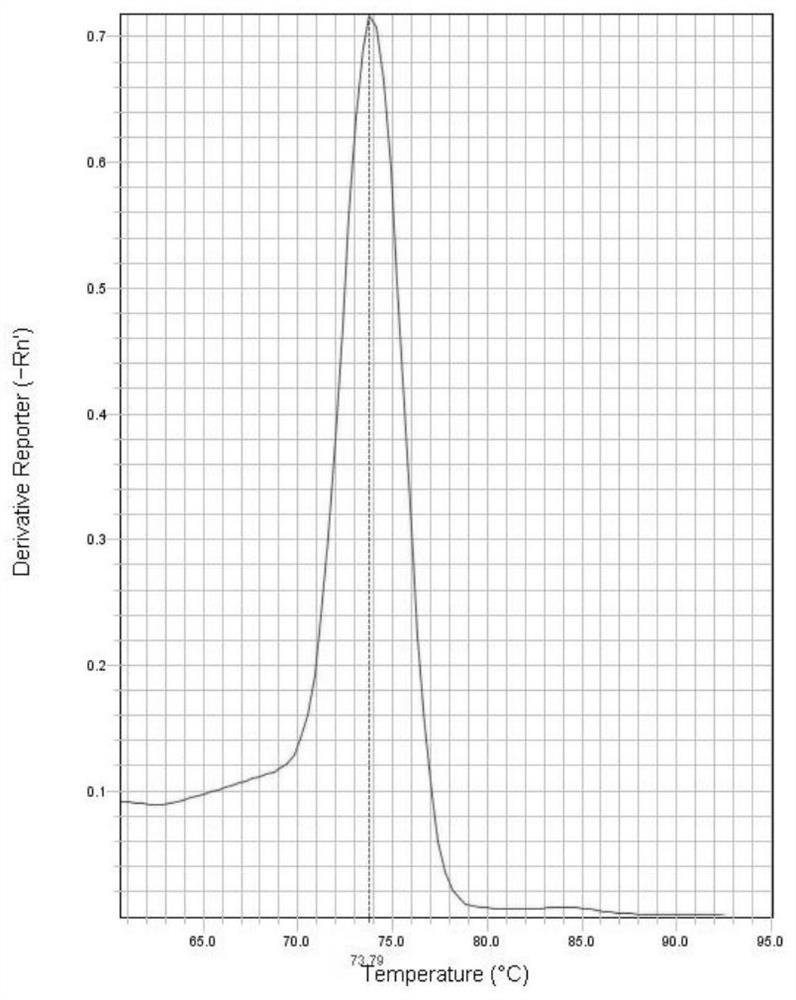
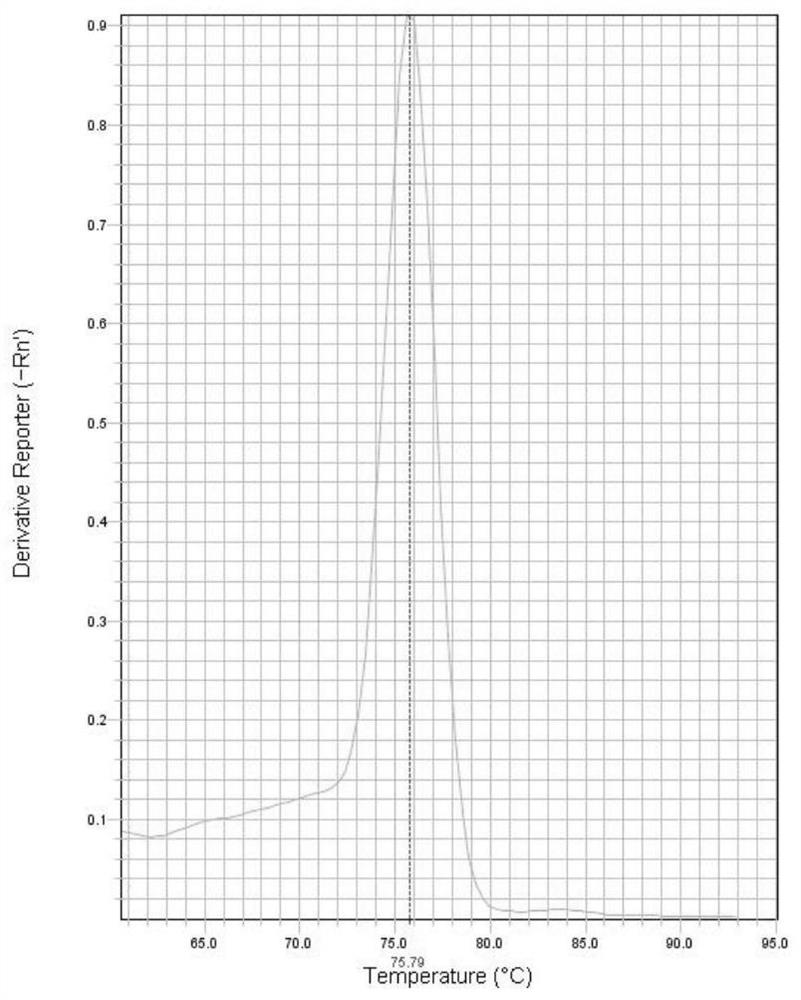
[0076] The amplification results showed that the melting curve of the LD126 gene ( figure 1 ) has a single peak at the Tm value of 73.79°C, and the melting curve of the LSDV001 gene ( figure 2 ) have only a single peak at Tm of 75.79°C. From the abov...
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com