CRISPR/Cas9 system causing congenital deafness and application of CRISPR/Cas9 system in preparation of model pig nuclear donor cells
A hereditary deafness, cell technology, applied to genetically modified cells, bone/connective tissue cells, cells modified by introducing foreign genetic material, etc., can solve the problems of unclear physiological effects and deafness-causing mechanism, and reduce work costs Quantity, good applicability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Embodiment 1, the preparation of plasmid
[0078] 1.1 Preparation of plasmid pU6gRNA eEF1a-mNLS-hSpCas9-EGFP-PURO (plasmid pKG-GE3 for short)
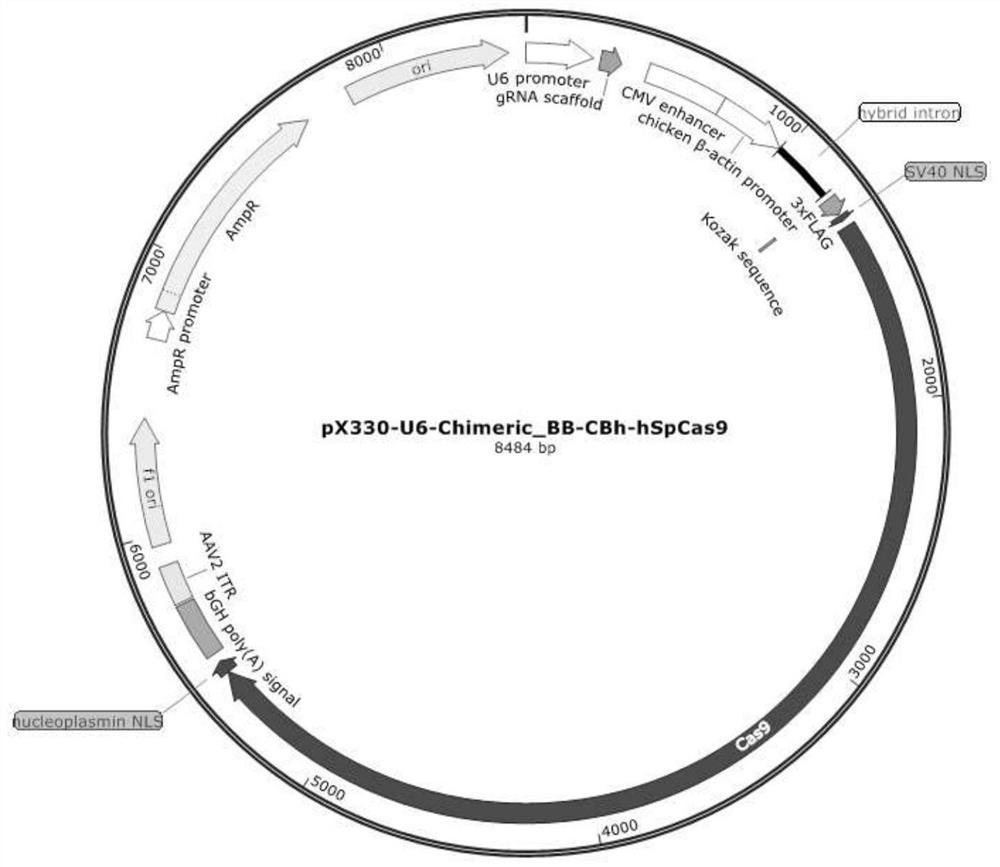
[0079] The original plasmid pX330-U6-Chimeric_BB-CBh-hSpCas9 (referred to as plasmid pX330, the sequence is shown in SEQ ID NO: 1. The structure diagram of plasmid pX330 is shown in figure 1 . In SEQ ID NO: 1, the 440-725 nucleotides form the CMV enhancer, the 727-1208 nucleotides form the chickenβ-actin promoter, and the 1304-1324 nucleotides encode the SV40 nuclear localization signal (NLS ), the 1325-5449th nucleotide encodes the Cas9 protein, and the 5450-5497th nucleotide encodes the nucleoplasmin nuclear localization signal (NLS).
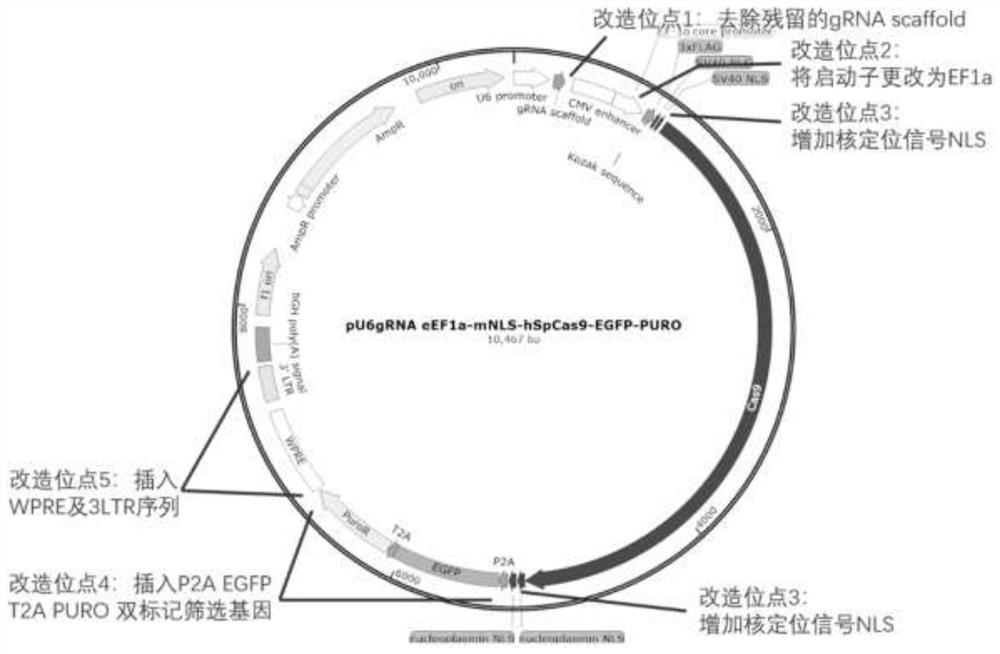
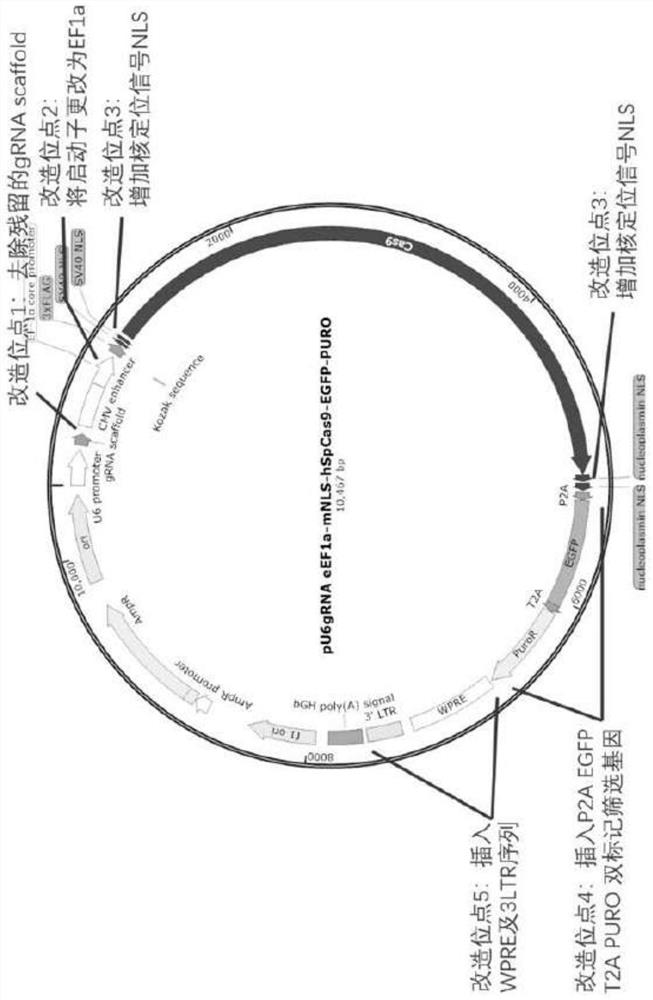
[0080] Plasmid pU6gRNA eEF1a-mNLS-hSpCas9-EGFP-PURO, referred to as plasmid pKG-GE3, the nucleotide is shown in SEQ ID NO:2. The build method is as follows:
[0081] (1) Remove redundant and invalid sequences in the gRNA backbone
[0082] Plasmid pX330 was digested with BbsI and XbaI, and ...
Embodiment 2
[0093] Embodiment 2, the effect comparison of plasmid pX330 and plasmid pKG-GE3
[0094] Select high-efficiency gRNA targets located at the RAG1 gene:
[0095] Target of RAG1-gRNA4: 5'-AGTTATGGCAGAACTCAGTG-3' (SEQ ID NO: 9).
[0096] Primers used to amplify and detect target-containing fragments are as follows:
[0097] RAG1-nF126: 5'-CCCCATCCAAAGTTTTTAAAGGA-3' (SEQ ID NO: 10);
[0098] RAG1-nR525: 5'-TGTGGCAGATGTCACAGTTTAGG-3' (SEQ ID NO: 11)
[0099] Primary pig fibroblasts were prepared from newborn Congjiangxiang pigs (female, blood type AO).
[0100] 1. Preparation of recombinant plasmids
[0101] The plasmid pKG-U6gRNA was taken, digested with restriction endonuclease BbsI, and the vector backbone (a large linear fragment of about 3 kb) was recovered. RAG1-4S and RAG1-4A were synthesized separately, then mixed and annealed to obtain double-stranded DNA molecules with cohesive ends. The double-stranded DNA molecule with cohesive ends and the vector backbone were lig...
Embodiment 3
[0123] Example 3, Target Screening for TMC1 Gene Knockout
[0124] Pig TMC1 gene information: encodes ransmembrane channel like 1 protein; located on pig chromosome 1;
[0125] GeneID is 100516949, Sus scrofa. The protein encoded by the porcine TMC1 gene is shown in GENBANK ACCESSION NO.XP_020920511.1 (linear MAM 13-MAY-2017). In genomic DNA, the porcine TMC1 gene has 27 exons, wherein the 12th exon and its upper and lower 500bp sequences are shown in SEQ ID NO: 14, and its encoded protein fragment is shown in SEQ ID NO: 15.
[0126] 1. Conservation analysis of predetermined targets of TMC1 gene knockout and adjacent genome sequences
[0127] 18 newborn Congjiang pigs, including 10 females (named 1, 2, 3, 4, 5, 6, 7, 8, 9, 10) and 8 males (named A, B, C, D , E, F, G, H).
[0128] Using the genomic DNA of 18 pigs as templates, PCR amplification was carried out using primer pairs (the target sequence of the primer pairs includes the 12th exon of porcine TMC1 gene), and then ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com