Bioactive molecule binding target identification method based on double-head photoaffinity probe
A bioactive molecule and identification method technology, which is applied in the field of bioactive molecule binding target identification based on double-headed photoaffinity probes, can solve the problems of increasing the number of potential bioactive small molecule binding proteins, difficulty in verification, and low efficiency. , to achieve good application prospects, excellent sensitivity and accuracy, and high purity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
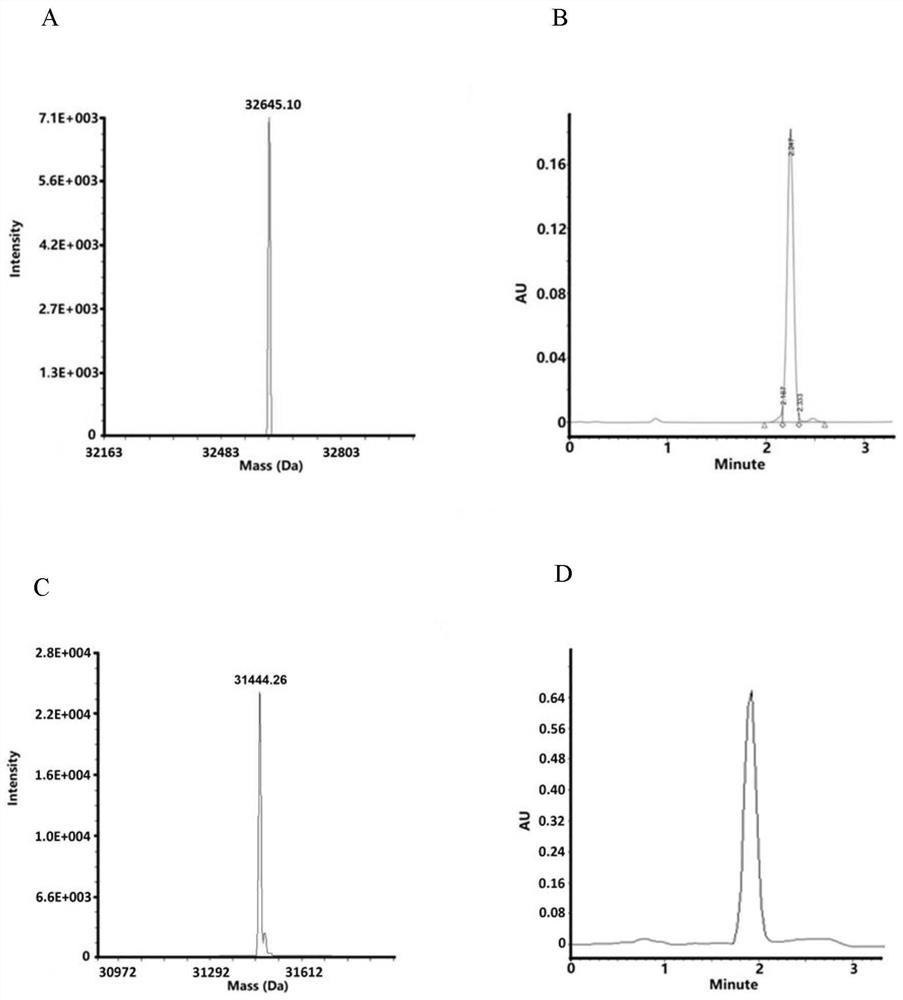
[0040] Example 1: Preparation, purification and characterization of double-headed photoaffinity probes
[0041] 1. Preparation of double-headed photoaffinity probes
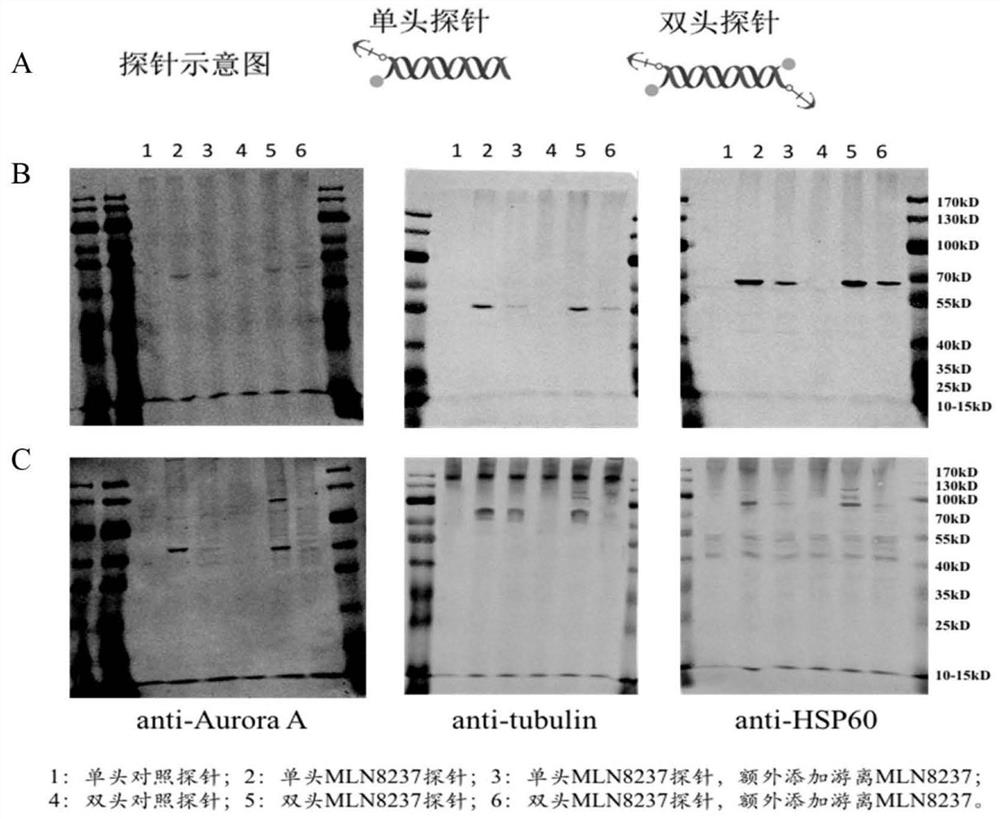
[0042] The double-headed photoaffinity probes are composed of double-headed binding probes and double-headed capture probes in groups. Double-ended binding probes are similar in structure to double-ended capture probes, see figure 2 A, Both include the probe framework in the middle and the active molecules and biological reaction amplification systems combined on both sides of the framework.
[0043] The probe backbone of the double-head binding probe binds the same biologically active molecules on both sides, the probe backbone of the double-head capture probe binds the same photoaffinity molecules on both sides, and the probe backbone of the double-head binding probe is the same as the double-head The probe backbone of the capture probe base-pairs or specifically binds.
[0044] In this embodiment, the prob...
Embodiment 2
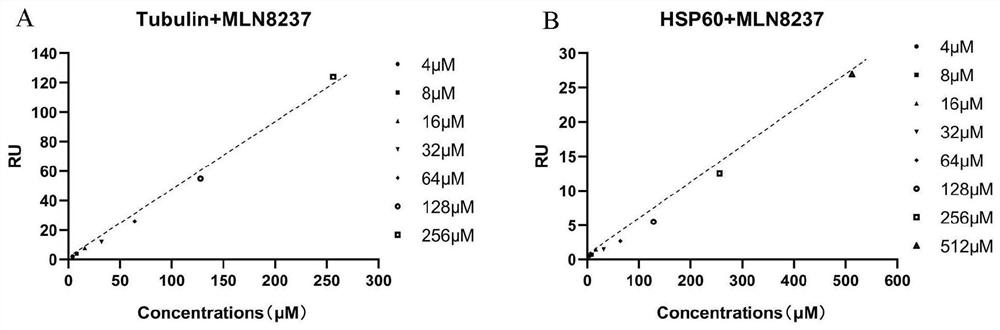
[0065] Embodiment 2 target identification and verification
[0066] 1. Extraction of whole-cell protein samples
[0067] (1) Cell culture and pretreatment:
[0068] Human hepatoma cell Hep3B was incubated at 37°C in 5% CO 2 In the cell culture incubator, culture in DMEM high-glucose medium containing 10% fetal bovine serum, 0.1mg / mL streptomycin, 100U / mL penicillin, and when the growth is about 80% confluent, use 0.25% trypsin to digest the cells, Add an appropriate amount of serum-containing medium to stop digestion, centrifuge at 1000g for 3min, discard the supernatant, resuspend in PBS, centrifuge at 1000g for 3min, and wash once.
[0069] (2) Protein extraction:
[0070] The above-mentioned cells were resuspended in western and IP cell lysates, placed in liquid nitrogen for 3 minutes, then placed in a water bath at 37°C for 5 minutes, repeated 3 times, centrifuged at 12000g for 5 minutes, and the supernatant was taken. Observe and compare the trypan blue staining under...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com