Breast tumor specific methylation detection kit
A detection kit and methylation technology, applied in biochemical equipment and methods, microbial determination/examination, DNA/RNA fragments, etc., can solve the problem of high sensitivity requirements for assay, achieve stable diagnostic ability, improve sensitivity and specificity sex, improve survival
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
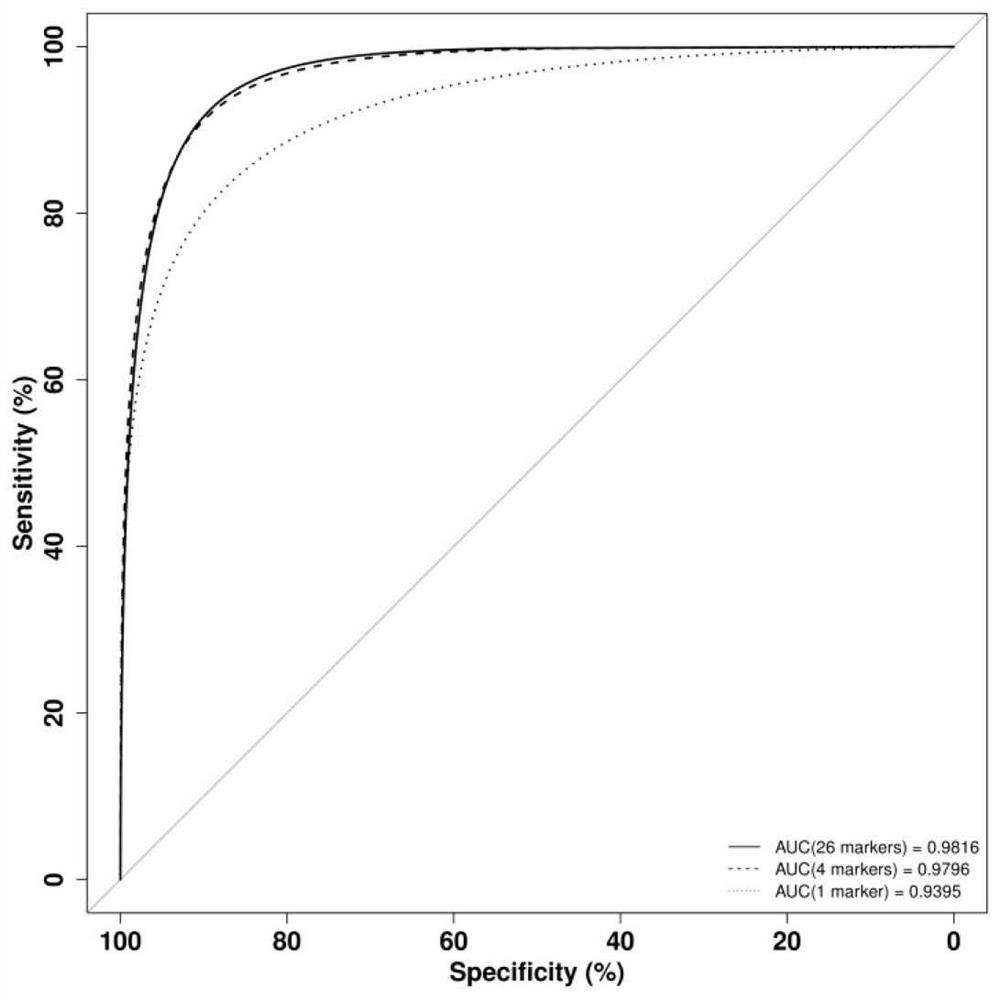
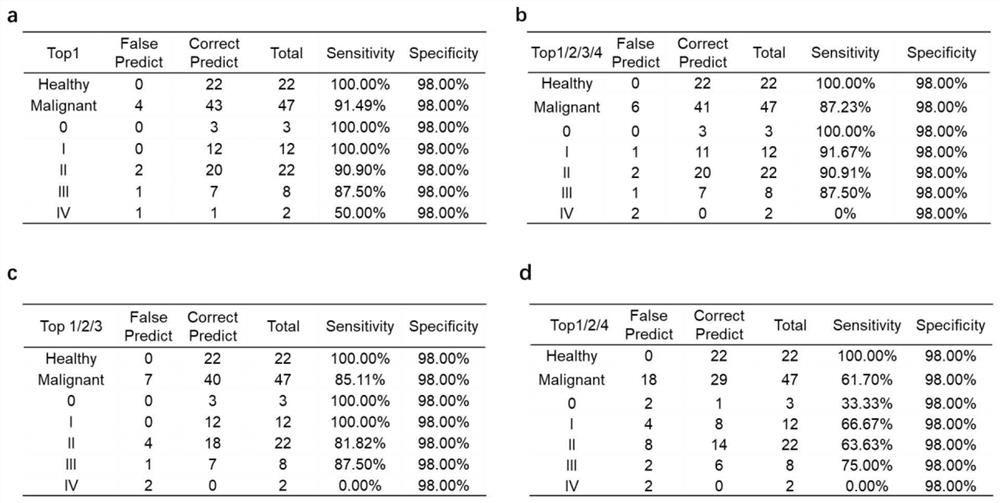
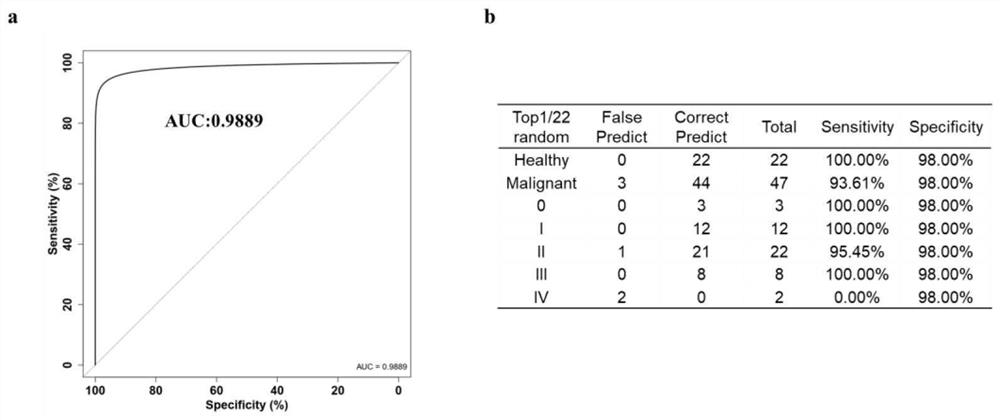
[0097] Example 1 This example provides a kit for detecting breast cancer in plasma samples, which contains 23 detection genes TLR5, C1orf61, NR2F1-AS1, ZDHHC1, OTP, FEZF2, TSHZ3, FLJ45513, POU3F1, NKX2- 1-DNA methylation molecular marker Marker1 of 23 methylated regions in AS1, PNPLA1, TIFAB, RADIL, EFCAB10, MAST1, ZFHX4-AS1, HOXB13, LHX5-AS1, LECT1, ALX3, BMP7, PRDM13 and ULBP1 Detection primers and probes to Marker23.
[0098] The kit has designed three pairs of primers and three probes for each of the specific methylation sites in the 23 molecular markers Marker1 to Marker23 for the detection of breast cancer in plasma samples (probe fluorescent labels can be FAM, VIC and NED and other fluorophore labels), and labeled as combinations 1, 2, and 3, respectively. Wherein, the combination of primers and probes selected in each molecular marker can be arbitrarily selected to be combined with combinations 1, 2, and 3 of primers and probes in other methylation markers and detecte...
Embodiment 2
[0105] In this example, the kit described in Example 1 was used to detect the methylation levels of 23 biomarkers in plasma samples.
[0106] A method for detecting methylation levels of DNA methylation molecular markers, comprising the following steps:
[0107] 1. Extraction of cfDNA from samples:
[0108] The specific operation steps of plasma DNA extraction were carried out according to the instructions of MagMAX™ Cell-Free DNA Isolation Kit of Life Company.
[0109] 2. Sulphite conversion of extracted cfDNA
[0110] The extracted cfDNA (10ng) was subjected to bisulfite conversion to deaminate the unmethylated cytosine in the DNA and convert it to uracil, while the methylated cytosine remained unchanged to obtain the bisulfite converted The specific operation of the DNA transformation was carried out according to the instructions of the EZ DNA Methylation-Lightning Kit of Zymo Research. Deamination of unmethylated cytosine in DNA to uracil, while methylated cytosine rema...
Embodiment 3
[0134] This embodiment provides the detection of molecular markers in standard products, and the detection steps are as follows:
[0135] 1. Standard product preparation
[0136] 1) Preparation of 0% methylation standard: using Single Cell Kit (Qiagen, Cat#150343) and Mung Bean Nuclease (NEB, Cat#M0250L) were used to process NA12878 DNA to prepare 0% methylation standard;
[0137] 2) Preparation of 100% methylation standard:
[0138] The prepared 0% methylation standard was treated with CpG Methyltransferase (M.SssI) to obtain a 100% methylation standard.
[0139] 2. Preparation of standard products with different methylation ratios:
[0140] Mix 0% and 100% methylation standards according to the desired methylation ratio gradient to obtain 0.2%, 0.4%, and 1% methylation ratio standards.
[0141] 3. Perform bisulfite conversion on standard DNA with different methylation ratios: the steps are as in Example 2, and the conversion input amount is 10-50 ng, preferably 50 ng in...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com