Application of high-throughput screening method using droplet micro-fluidic chip in actinomycetes
A microfluidic chip and screening method technology, applied in the field of microbial biology, can solve the problem of high-throughput screening and detection without actinomycetes, and achieve the effect of wide detection and sorting time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
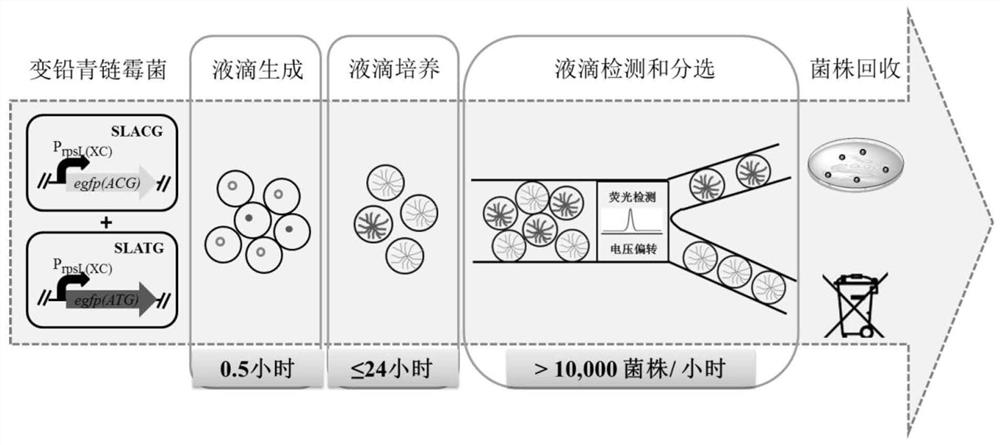
[0039] Example 1 Construction of negative and positive strains with green fluorescent signal
[0040] On the basis of the sequence of the original enhanced green fluorescent protein gene egfp (as described in SEQ NO.1 in the sequence listing), ATG is used as the start codon, and all GTG codons in the expression frame are synonymously mutated to GTC or GTA to obtain the egfp (ATG) gene (as described in SEQ NO.2 in the sequence listing). On the basis of egfp(ATG), the initial ATG was replaced with ACG by point mutation method to obtain the egfp(ACG) gene (as described in SEQ NO.3 in the sequence listing). Select 5 heterologous promoters (Table 1): P gapdh(EL) , P rpsL(XC) , P rpsL(SG) , P rpsL(TP) , P erm*E , synthesize these promoter sequences, and use the synthetic sequences as templates to clone these promoters respectively; select 4 endogenous promoters at the same time (Table 1): P rpS12 , P gpmA , P pyk , P rpoA , and these promoters were cloned using the Streptomyc...
Embodiment 2
[0058] Embodiment two plate spore culture and collection
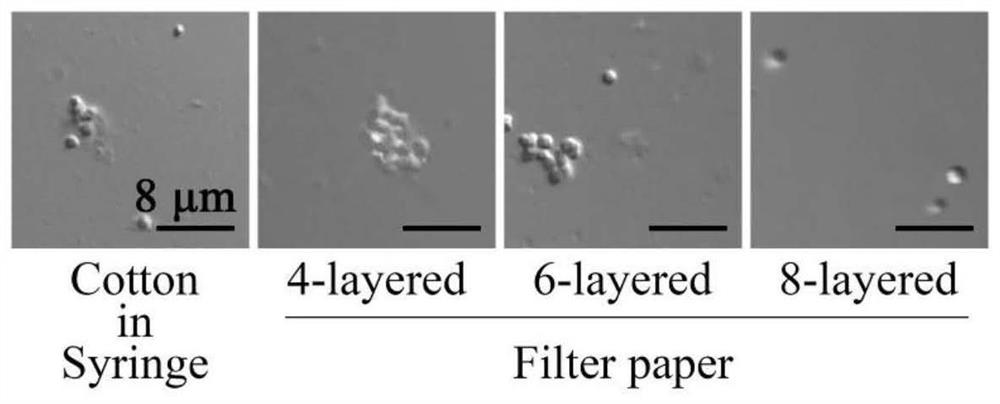
[0059] Take the spores (suspended in 20% glycerol solution) frozen at -80°C, dip a small amount of spore liquid with a sterile inoculation loop, and streak and passage on the R2YE solid plate. Plates were placed upside down in a 30°C incubator and cultured for 5-7 days until gray mature spores differentiated. When collecting, add 5 mL of sterile R2YE liquid medium that has passed through a 0.22 μm aqueous membrane to the surface of the plate, and repeatedly blow and suck the spores on the plate with a pipette to suspend them in the medium. Take 3 mL of the eluted spore suspension and add it to a sterile 8-layer lens paper filter device, and filter twice until the spores are in a monodisperse state in the medium (such as figure 2 shown). Take 20-30 μL of the filtered spore suspension and drop it on a hemocytometer, observe and count the spore concentration under a 20x objective lens. Dilute the spore suspension with...
Embodiment 3
[0060] Embodiment three droplet embedding method
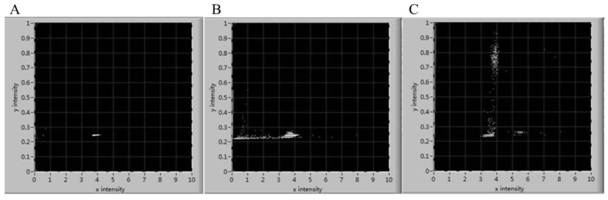
[0061] Droplet embedding according to methods in published literature He, R., Ding, R., Heyman, J.A. et al. JInd Microbiol Biotechnol (2019) 46:1603. https: / / doi.org / 10.1007 / s10295- 019-02221-2: Wherein the water phase is 1mL and the spore concentration is 1×10 6 spore suspension per mL, add the above water phase into a 1mL syringe, the oil phase is 1mL oil containing stabilizer, also add into a 1mL syringe, and then the syringe with the water phase and the oil phase and the droplet form Microchip Unicom, try to adjust the ratio of the flow rate of the oil phase and the water phase to 1:1, 2:1 and 3:1, respectively, to generate droplets of different diameters ( Figure 3-B , C). When the ratio of spore concentration to the number of droplets is between 0.3-0.5 and the diameter of the obtained droplets is 80-100 μm, the formation ratio and detection speed of spore droplets are optimal. The size of the prepared droplet is rel...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com