Antigen epitope peptide and application thereof of a kind of novel coronavirus T cell
An antigenic epitope and coronavirus technology, applied in the direction of virus antigen components, viral peptides, viruses, etc., can solve the problem of no research reports on the new coronavirus
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
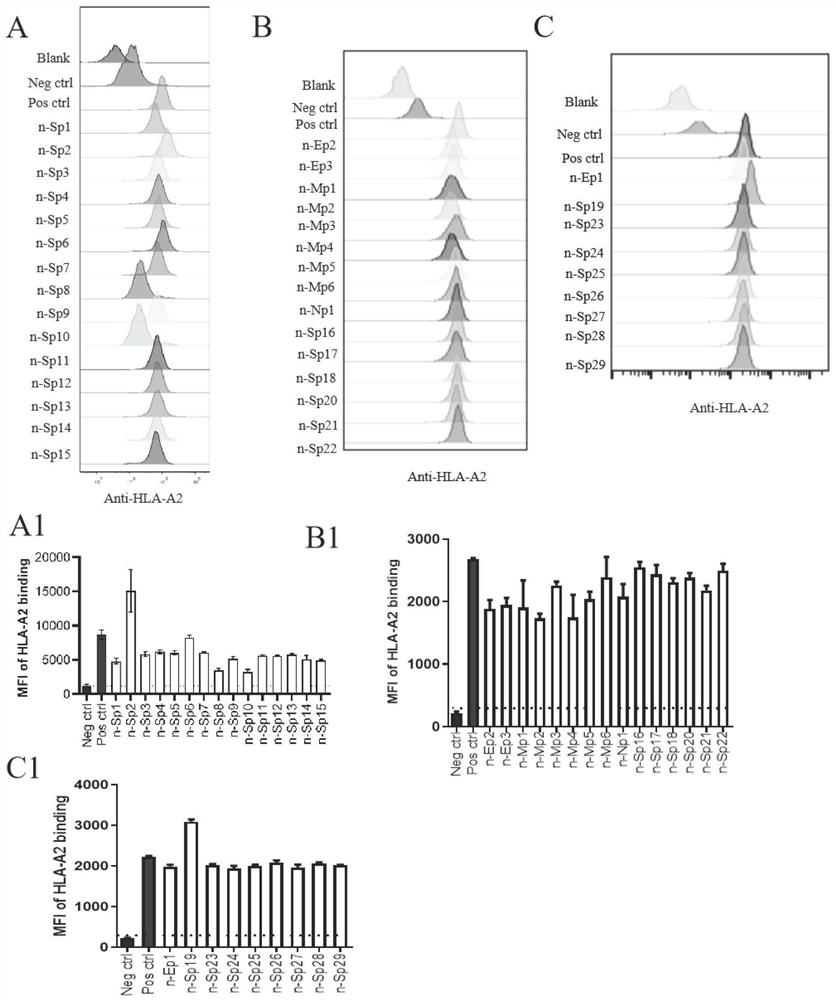
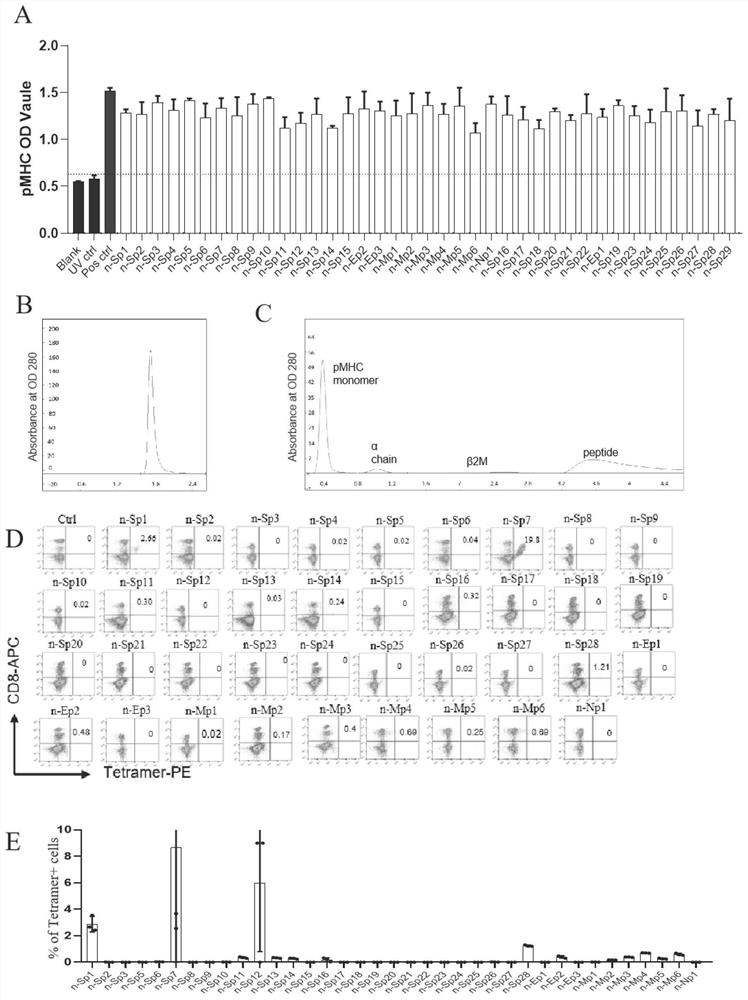
[0058] Example 1 Prediction of New Coronavirus HLA-A2 Restricted Epitopes
[0059] 1. Experimental method
[0060] Using the MHC-I binding tool (http: / / tools.iedb.org / mhci), SARS-CoV-2 Wuhan strain (NC_045512.2), SARS-CoV-GD01 strain (AY278489.2), MERS-CoV strain (KF600612.1) and human coronavirus OC43 strain (KF530099.1) spike (S), membrane (M) and nucleocapsid (N) protein sequences for T cell epitope prediction. The prediction method used was 2.22 (NetMHCpan-EL) recommended by IEDB, and the MHC allele selection was HLA-A*02:01 and HLA-A*02:06 (the most common HLA class I genotypes in the Chinese population) . Variant sequences for each epitope were extracted from the GISAID database (GISAID.org website) for further analysis.
[0061] 2. Experimental results
[0062] A large number of high-efficiency and specific T cell antigen epitope candidates were screened. Its information is shown in Table 1.
[0063] Table 1 Novel coronavirus T cell antigen epitope peptide:
[00...
Embodiment 2
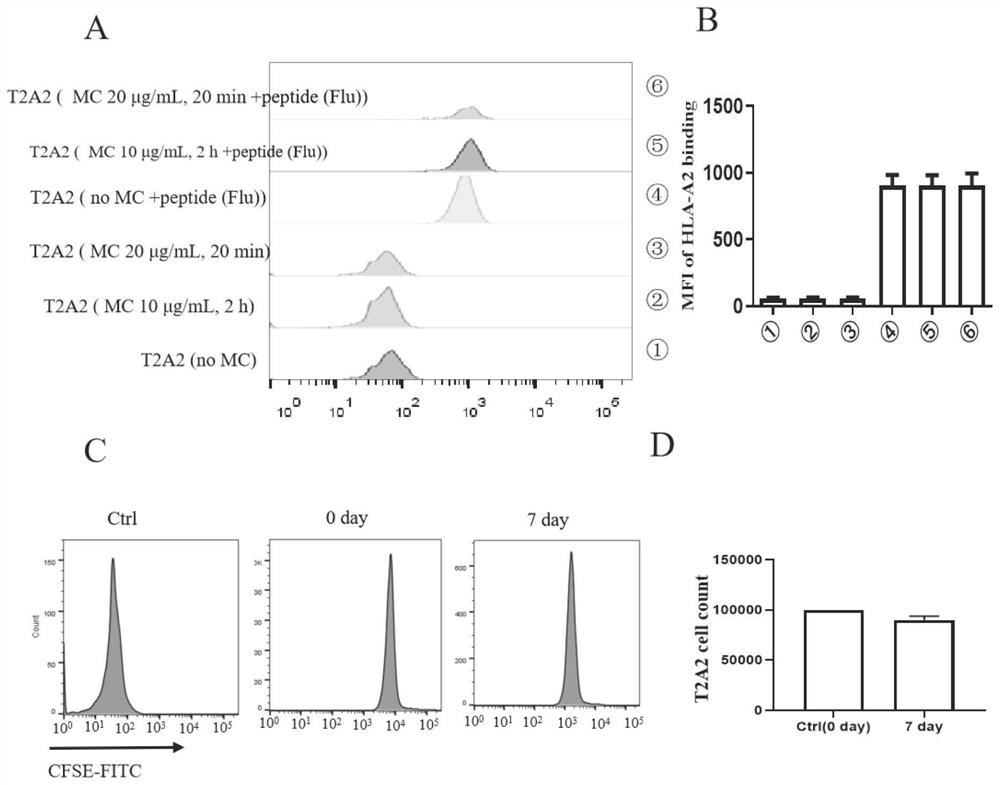
[0066] Example 2 Does Mitomycin Intervention T2A2 Cells Affect the Combination with Antigen Peptides
[0067] 1. Experimental method
[0068] Take logarithmic growth state T2A2 cells (174x CEM.T2( CRL-1992 TM )), seeded into 96-well plate, 10 per well 5 , select different concentrations of mitomycin (MC) (20μg / mL and 10μg / mL) and different action time (20min and 2h) to intervene in T2A2 respectively, set up 6 groups, respectively, group 1: T2A2 blank group, group 2 : T2A2+MC 10μg / mL 2h, group 3: T2A2+MC 20μg / mL 20min, group 4: T2A2+10μM positive control peptide (influenza A M1 polypeptide GILGFVFTL), group 5: T2A2+MC 10μg / mL 2h+10μM positive Control peptide (influenza A M1 polypeptide GILGFVFTL), group 6: T2A2+MC 20 μg / mL 20 min+10 μM positive control peptide (influenza A M1 polypeptide GILGFVFTL). Each group has 3 replicate wells with a final volume of 200 μL. After incubation at 37°C for 4 hours, centrifuge and wash twice, label with FITC anti-human HLA-A2 (β2m) antibo...
Embodiment 3
[0071] Example 3 Mitomycin inhibits T2A2 cell proliferation
[0072] 1. Experimental method
[0073] Take T2A2 cells in the logarithmic growth state and plant them into 96-well plates, 10 per well 5 , take mitomycin (MC 20μg / mL), the action time (20min) to intervene in T2A2, set up 3 groups, respectively T2A2 blank group, label T2A2 with CFSE (5μM) (day 0), and label T2A2 with CFSE (5μM) (Day 7), each group had three replicate wells, and the final volume was 200 μL. Placed at 37°C, 5% CO 2 The cells were cultured in the incubator, centrifuged and washed twice after the corresponding time, and detected by flow cytometry. The experiment was carried out 3 times.
[0074] 2. Experimental results
[0075] After mitomycin MC intervened in T2A2, it could inhibit the cell proliferation ( figure 1 C and panel D). figure 1 C shows that CFSE labeling was successful (the fluorescence peak of the middle group shifted to the right on day 0). After the 7th day, CFSE still showed a si...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com