Neutralizing antibody against novel coronavirus receptor binding regions and application thereof
A coronavirus, receptor binding technology, applied in the direction of antiviral agents, viruses/phages, antiviral immunoglobulins, etc., to achieve the effect of improving the neutralization ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 110
[0105] Example 1 10×Genomics database construction and data analysis
[0106] In this example, the peripheral blood mononuclear cells of patients with COVID-19 were first extracted by lymphocyte separation medium, and CD19 阳性 CD27 阳性 Memory B cells and CD19 阳性 CD27 高 CD38 高 Plasma cells, and label B cells specific to the receptor binding region of the new coronavirus;
[0107] The prepared cells were subjected to 10× Genetics library construction and sequencing, and three libraries were constructed: 5' transcriptome library, B cell receptor library and signature library, and finally high-throughput sequencing was performed using NovaSeq 6000;
[0108] Use 10×Genomics Cell Ranger software (version 3.1.0) to process sequencing fastq files, use Seurat (version 3.1) to filter low-quality cells (gene expression 5000, mitochondrial genes>10%, gene expression less than 3 cells), and finally use Seurat for standardization, PCA dimensionality reduction, TSNE clustering, and differ...
Embodiment 2
[0109] Example 2 Synthesis and expression of antibody
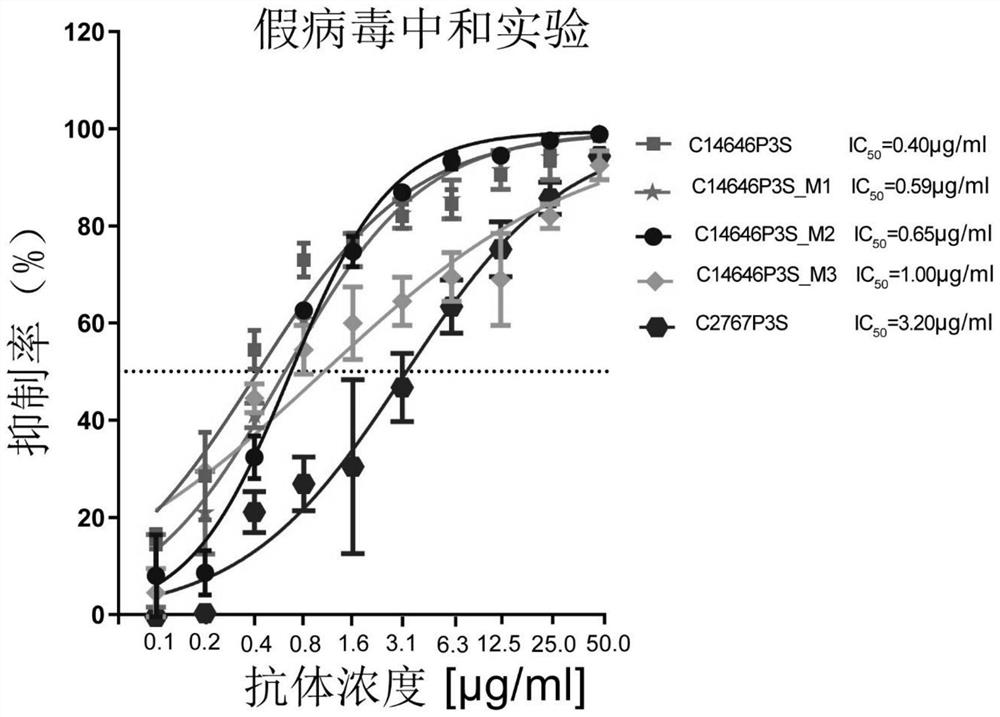
[0110] Candidate antibodies were selected according to the following principles: 1) antibody clonal types other than IgE and IgD, 2) clones with high and independent antigen labeling, 3) clones with high somatic mutation rate excluded, 4) clones with high-frequency V-J combinations. Antibody C14646P3S (SEQ ID NOs: 7-8) and antibody C2767P3S (SEQ ID NOs: 17-18) were obtained from this screening.
[0111] The heavy chain CDR3 of C14646P3S was further mutated to obtain neutralizing antibody mutants C14646P3S_M1 (SEQ ID NO:22, SEQ ID NO:8), C14646P3S_M2 (SEQ ID NO:25, SEQ ID NO:8), C14646P3S_M3 (SEQ ID NO: 28, SEQ ID NO: 8).
[0112] The screened antibody gene was linked to an IgG carrier containing a constant region, and the antibody was expressed by transfecting HEK293T cells, adding a transfection reagent, collecting the supernatant and performing antibody purification.
Embodiment 3
[0113] Specific identification of embodiment 3 antibody
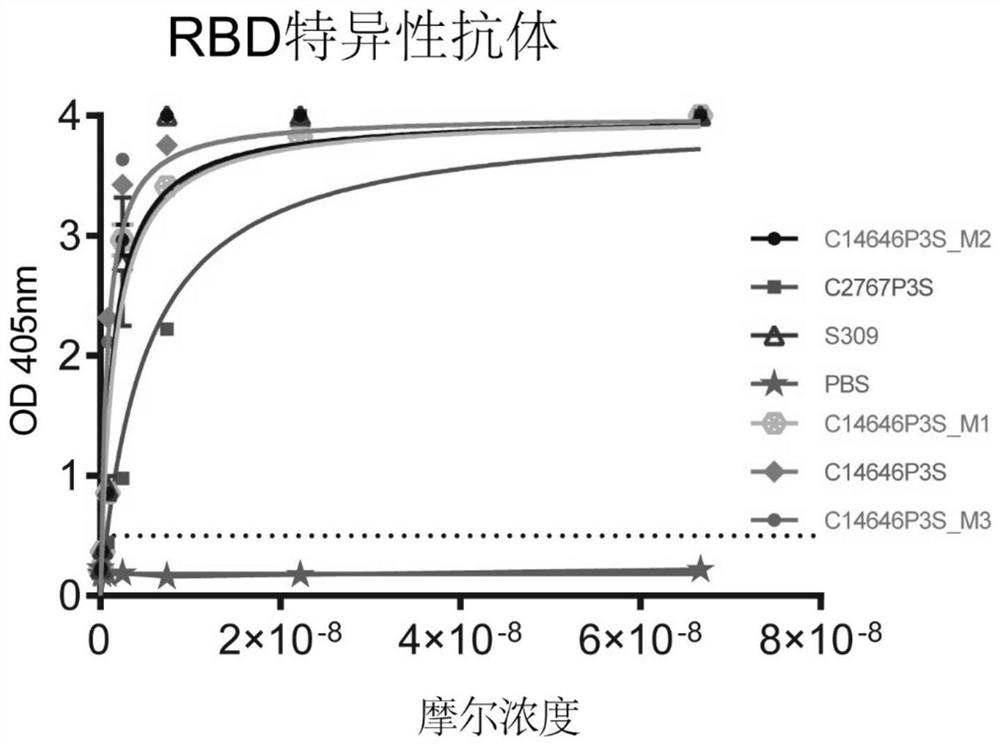
[0114] This embodiment utilizes ELISA to identify the specificity of the antibody, prepares a microtiter plate coated with the SARS-CoV-2 receptor binding region antigen, and adds screening antibody C14646P3S and its mutant antibody C14646P3S_M1, C14646P3S_M2, C14646P3S_M3, C2767P3S or positive control antibody (S309 ), followed by the addition of HRP-labeled goat anti-human IgG secondary antibody for ELISA detection.
[0115] The result is as figure 1As shown, C14646P3S and its mutant antibodies C14646P3S_M1, C14646P3S_M2, C14646P3S_M3 and C2767P3S monoclonal antibodies strongly bind to the receptor binding region.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com