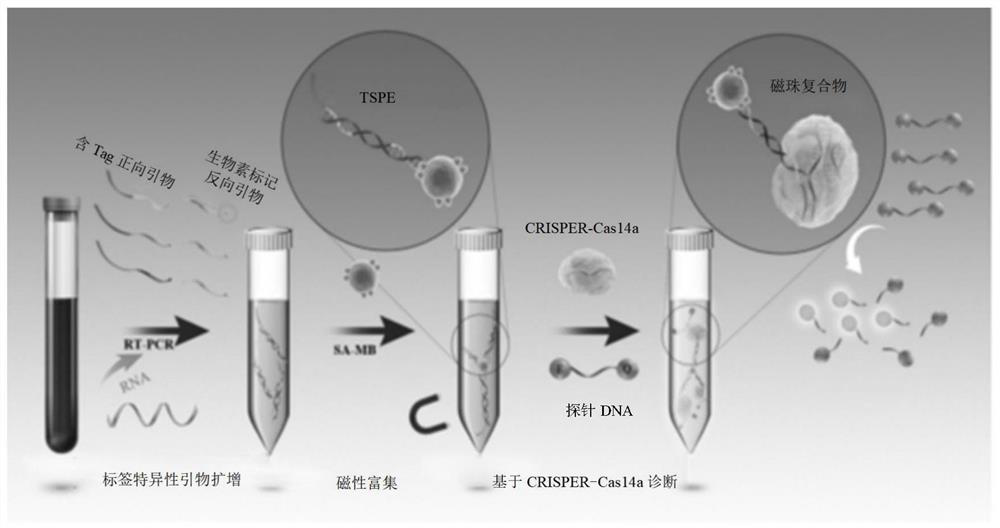
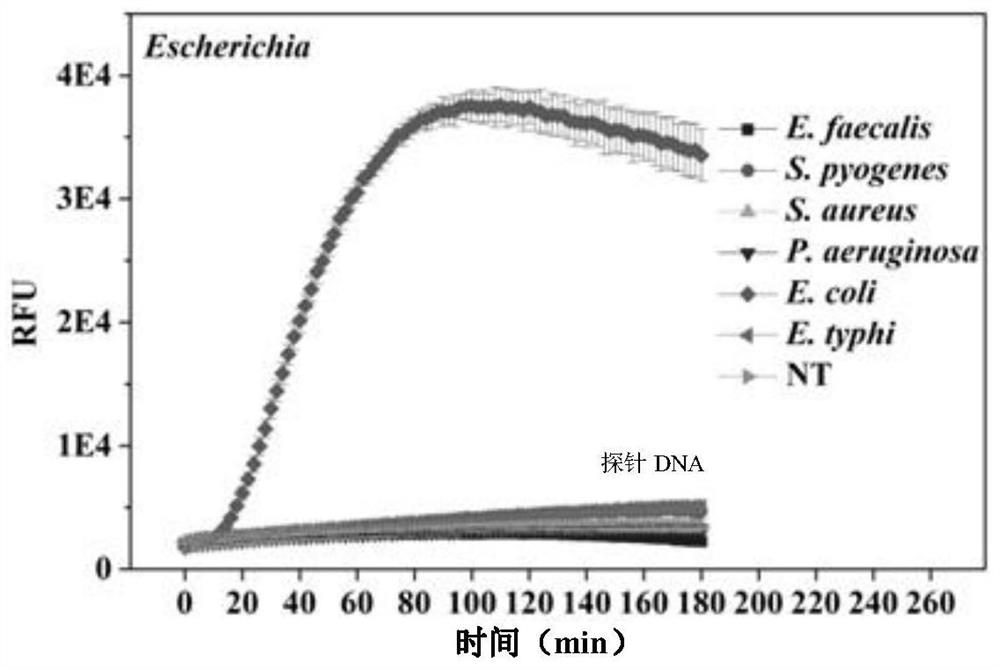
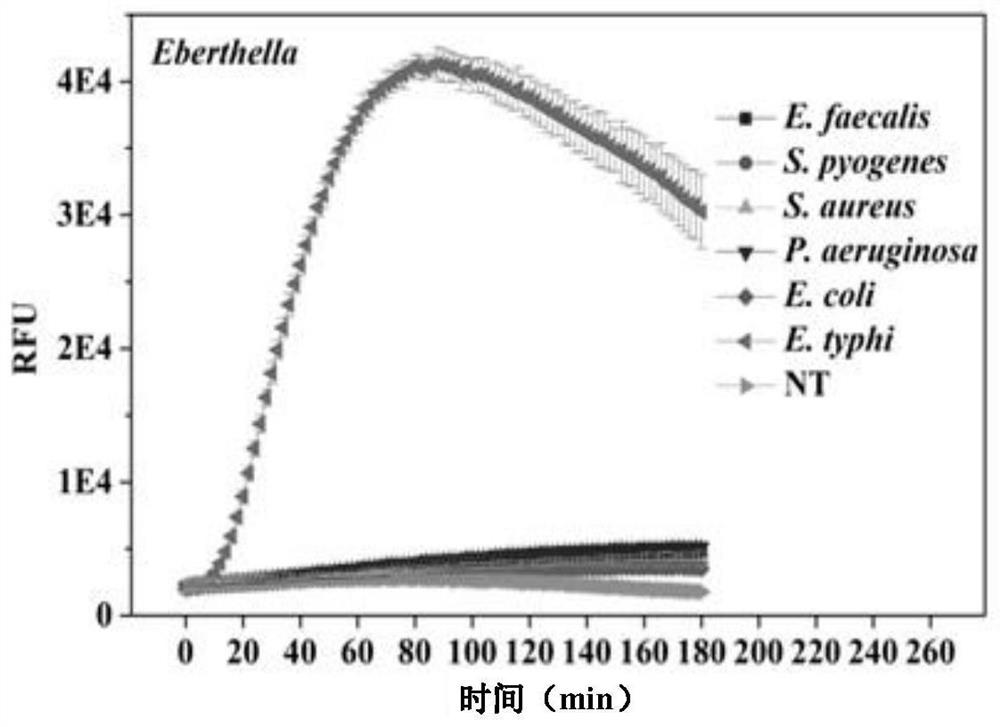
General bacterium and virus detection system and method based on CRISPR-Cas14a and application
A technology for virus detection and bacteria, applied in the field of general bacteria and virus detection systems, can solve the problems of increasing detection difficulty and detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0041] (2) Preparation of general sgRNA
[0042]A template for universal sgRNA was amplified by PCR from the pUC19 vector containing the T7 promoter, the nucleotide target sequence (SEQ ID NO.1) matching the sequence shown in Table 1, and sgRNA resistance. Use the amplified PCR product as a DNA template, transcribe the sgRNA in vitro at 37°C for 16 hours with the T7 Rapid High-yield RNA Synthesis Kit, and then use the RNA Purification Kit (NEB) to purify to obtain a universal sgRNA and store it at -70°C for future use .
[0043] Table 1 Sequence of universal sgRNA
[0044]
preparation example
[0046] (1) Culture of Escherichia coli and preparation of cDNA
[0047] Escherichia coli was purchased from China Industrial Culture Collection Center (CICC) and cultured according to the manufacturer's instructions. That is, use LB medium (10g / L trypsin, 5g / L yeast extract and 10g / L sodium chloride) to inoculate Escherichia coli, keep a constant temperature at 37°C, and cultivate overnight at 180rpm, until it grows to 10 8 After CFU / mL, the bacteria were centrifuged at 10000rpm for 10 minutes, and the bacterial RNA kit (E.Z.N.A. Bacterial RNA Kit, OMEGA) was used to extract bacterial RNA.
[0048] The bacterial RNA extracted above was subjected to a thermal cycle condition of 42°C for 60 minutes using the PrimeScript II first-strand cDNA synthesis kit (Takara), and then subjected to a thermal cycle condition of 70°C for 5 minutes, and finally, E. coli was obtained. cDNA.
[0049] (2) Culture of Salmonella typhi and preparation of cDNA
[0050] Salmonella typhi was purcha...
Embodiment 1
[0067] (1) Preparation of Escherichia coli TSPE
[0068] The target region (SEQ ID NO.2) of E. coli was selected by MegAlign software (DNASTAR). Primers were designed by Premier software according to the target region, wherein the forward primer (SEQ ID NO.3) was combined with a Tag tag, and the reverse primer (SEQ ID NO.4) was combined with a biotin tag. The sequences are shown in Table 2.
[0069] 200nM Escherichia coli-specific forward and reverse primers (wherein the forward primer has a Tag sequence and the reverse primer has a biotin sequence) and six kinds of bacteria such as Escherichia coli and the cDNA of SARS-CoV-2 are used as templates respectively, Add DreamTaq TM Hot-start DNA polymerase (Thermo) adopts traditional PCR technology to carry out PCR amplification to prepare TSPE products respectively.
[0070] The PCR system is: DreamTaq TM Hot-start DNA polymerase 1 μL, 10X buffer 5 μL, 10 mM dNTPs 1 μL, upstream and downstream primers (forward and reverse prim...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com