Novel coronavirus mutant strain S protein and subunit vaccine thereof
A subunit vaccine, coronavirus technology, applied in the field of new coronavirus mutant S protein and its subunit vaccine, can solve the problems of decreased neutralization effect and low effectiveness of B.1.351
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0062] According to another typical implementation of the embodiments of the present invention, a method for preparing the S protein of a novel coronavirus mutant strain is provided, the method comprising:
[0063] obtaining the recombinant expression vector;
[0064] Transfecting the recombinant expression vector into cells, and obtaining a cell line stably expressing the recombinant S protein through glutamine resistance screening and monoclonal screening of the cell population;
[0065] The cell line was secreted, expressed and purified to obtain a purified recombinant novel coronavirus mutant strain S protein.
[0066] According to another typical embodiment of the embodiment of the present invention, a subunit vaccine of a novel coronavirus mutant strain is provided, and the novel coronavirus mutant strain subunit vaccine comprises the recombinant S protein and a pharmaceutically acceptable adjuvant.
[0067] The adjuvant includes at least one of aluminum hydroxide, leci...
Embodiment 1
[0070] Example 1. Prediction of Consensus Sequence
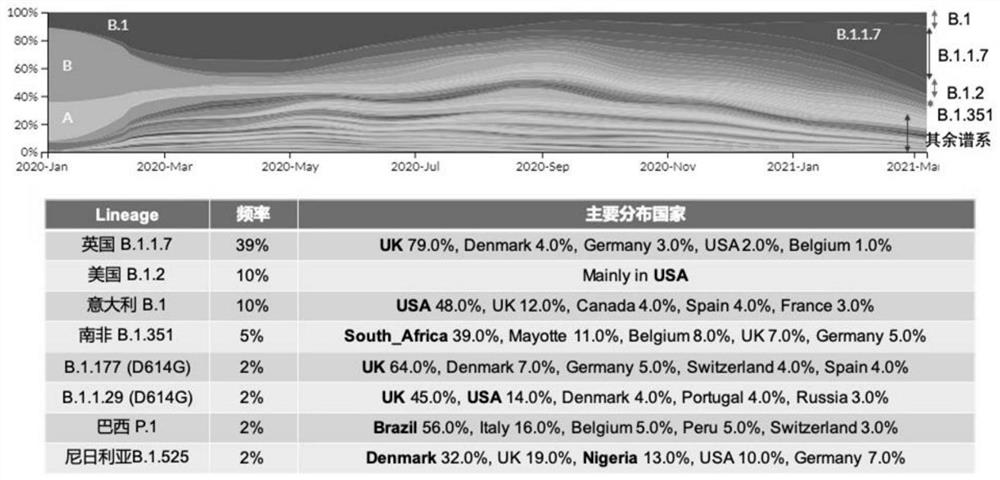
[0071] Due to the high mutation of SARS-CoV-2, especially the mutation site in the key receptor-binding domain (RBD) on the S protein, it may strengthen its ability to bind to the receptor ACE2, increasing the transmission or pathogenicity of the virus At the same time, the mutation may change the key epitope of the antigen, reduce the affinity of the neutralizing antibody that has been screened, or reduce the protective effect of the vaccine and neutralizing antibody. Currently, SARS-CoV-2 has evolved various types of mutations ( figure 1 ). For these mutants, we adopted four different strategies to predict and compare the consensus sequences of the mutants by software.
[0072] 1. Strategy 1:
[0073]We downloaded 2674 (deduplicated) S protein sequences of 2019-nCoV from the NCBI database (as of February 28, 2021), and constructed a phylogenetic tree of these S proteins by the neighbor-joining method using MEGA 7.0 softw...
Embodiment 2
[0082] Through four strategies, we obtained the S protein consensus sequence (S6) with 6 typical mutations (69-HV-70 deletion, Y144 deletion, E484K, N501Y, D614G, P681H) and 15 characteristic mutations (69- HV-70 deletion, Y144 deletion, 242-LAL-244 deletion, and contains the S protein consensus sequence (S15) of A222V, K417N, L452R, E484K, N501Y, A570D, D614G, Q677H, P681H, T716I, S982A, D1118H). Example 2. Construction and expression optimization of recombinant S protein vector
[0083] 1. Construction of S protein gene expressed in mammalian cell supernatant
[0084] The schematic diagram of the construction of the S protein expression gene of the present invention is shown in Figure 6 and Figure 7 .
[0085] Figure 6 : Diagram of the consensus sequence S6 schema. The sequence contains deletions of 69-70 and 144 amino acids, and contains E484K, N501Y, D614G, P681H mutations. At the same time, in order to maintain the integrity of the recombinant S protein, the Furi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com