Chitobiose deacetylase mutant and application thereof
A technology of deacetylase and chitobiose, applied in the field of bioengineering, can solve the problem of low efficiency of glucosamine GlcN and achieve the effect of increasing the expression level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Molecular docking and site selection
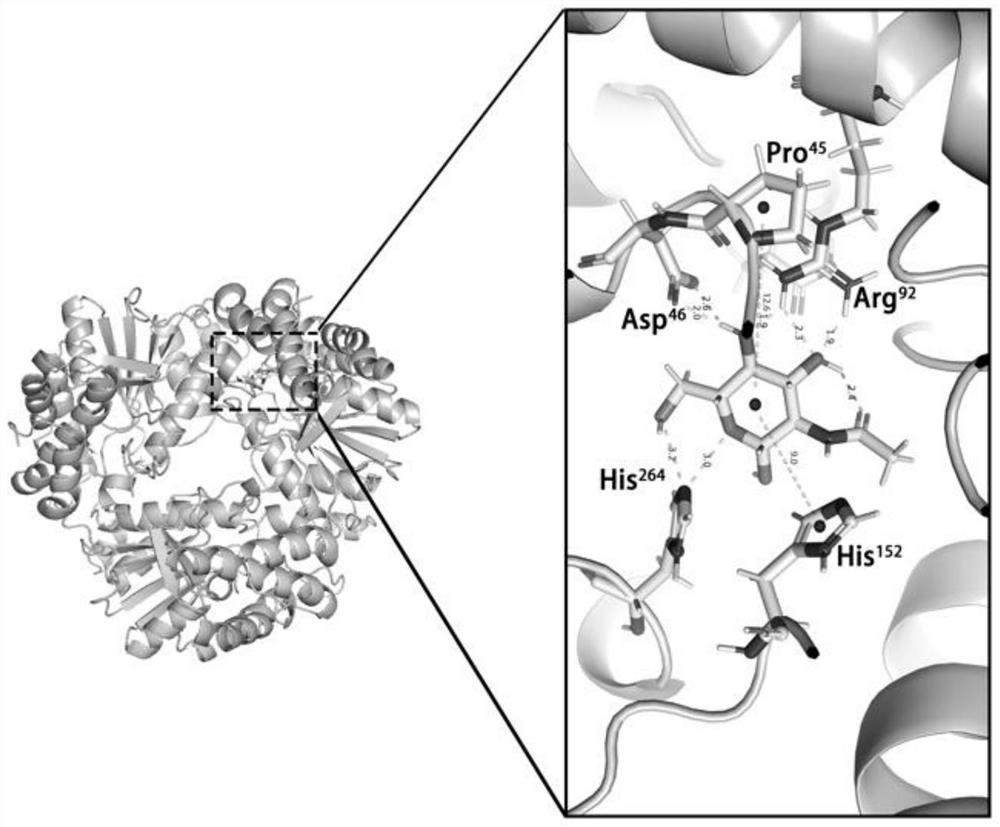
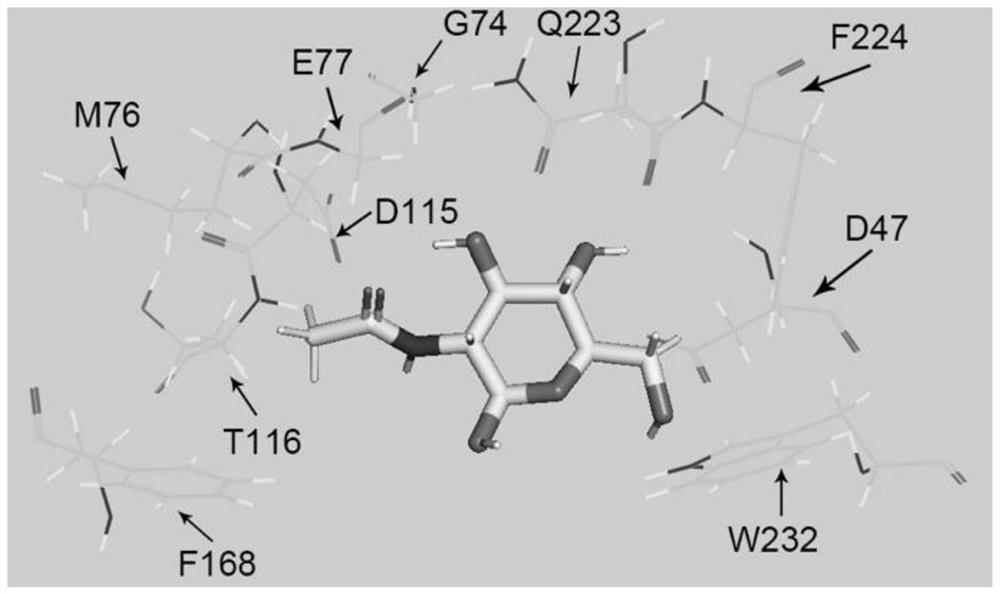
[0034] In order to improve the catalytic efficiency of Dac, the CDOCKER module of the software Discovery Studio was used to carry out molecular docking experiments to determine the binding site between the substrate molecule and the protein molecule and the key sites that affect the catalytic efficiency of the protein, so as to select the amino acid residues for alanine scanning base. Taking the three-dimensional structure model of Dac as the molecular protein and the substrate GlcNAc as the ligand, figure 1 The five sites where Dac is closest to the substrate and can form hydrogen bonds figure 2 10 sites that are slightly farther away from the substrate According to the molecular docking results, 15 sites were selected for alanine scanning, namely P45, D46, D47, G74, M76, E77, R92, D115, T116, H152, F168, Q223, F224, W232, H264.
Embodiment 2
[0036] Mutant construction of predicted loci
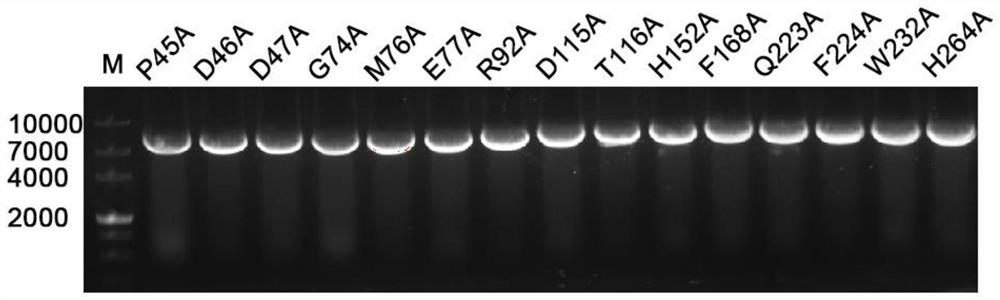
[0037] According to the base sequence of chitobiose deacetylase Dac, degenerate primers were designed, as shown in Table 1. The target gene (sequence shown in SEQ ID NO.1) was inserted into the expression vector pP43NMK (restriction site is KpnI and PstI) by double restriction ligation to construct recombinant plasmid pP43NMK-Dac. Design site-directed mutagenesis primers according to the coding sequence of Dac, as shown in Table 1, carry out PCR amplification (such as image 3 ), and E. coli Escherichia coli JM109 transformation, after the plasmid was extracted, it was transformed into the expression host Bacillus subtilis WB600.
[0038] Table 1 Site-directed mutagenesis primer sequences
[0039]
[0040]
Embodiment 3
[0042] Mutant extracellular enzyme activity assay
[0043]The constructed monoclonal transformants of 15 mutant strains were picked into 50 mL centrifuge tubes containing 5 mL liquid LB medium for seed culture, and 10 mg / mL kanamycin was added to each tube. The seed solution was cultured on a spring shaker at 37°C for 12 hours, and then transferred to a 500mL Erlenmeyer flask containing 96mL liquid TB medium with a 4% inoculum size for fermentation, and 10mg / mL kanamycin was added to each bottle. The enzyme activity of the crude enzyme solution is detected by the phthalaldehyde chromogenic method, and the specific method is as follows:
[0044] After 60 hours of fermentation, 1 mL of the fermentation broth was taken and centrifuged at 12000 rpm at 4°C for 2 minutes. The supernatant is the crude enzyme solution, and the substrate is 100 g / L N-acetylglucosamine GlcNAc solution prepared with pH 8.0 phosphate buffer. Put 100 μL each of the crude enzyme solution to be tested and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com