Kit for extracting genome DNA based on magnetic material and application method
A magnetic material and genome technology, applied in the field of genomic DNA extraction, can solve the problems of complicated preparation process of magnetic beads and poor DNA extraction effect, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
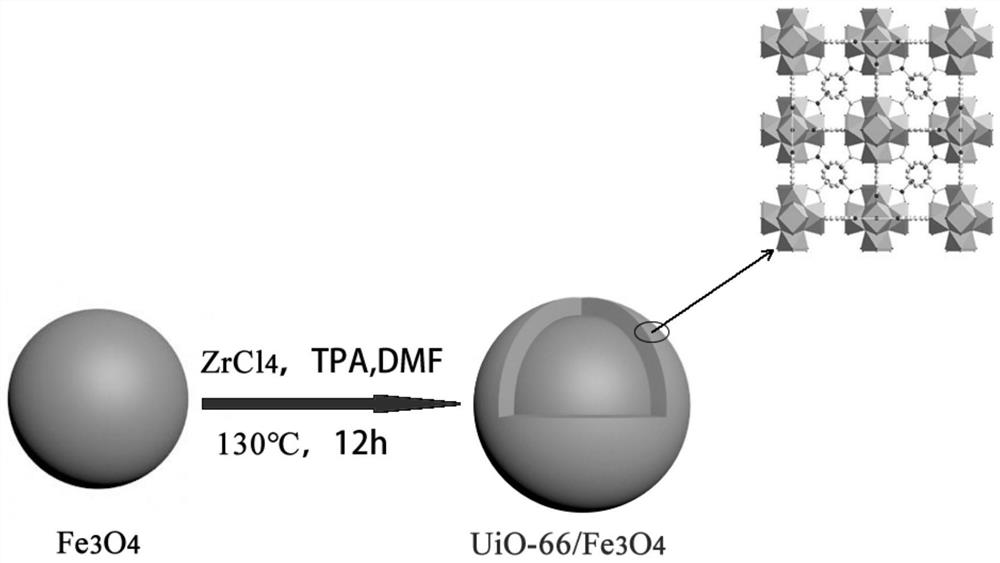
[0046] A kit for extracting bacterial genomic DNA based on magnetic materials, including lysate, binding solution, washing solution, eluent and magnetic beads, the magnetic beads are Fe 3 o 4 @UiO-66 composite material, the eluent is TE buffer; the washing solution includes the first washing solution and the second washing solution.
[0047] Specifically, the magnetic beads, lysate, first washing solution, second washing solution, and eluent are prepared by the following methods:
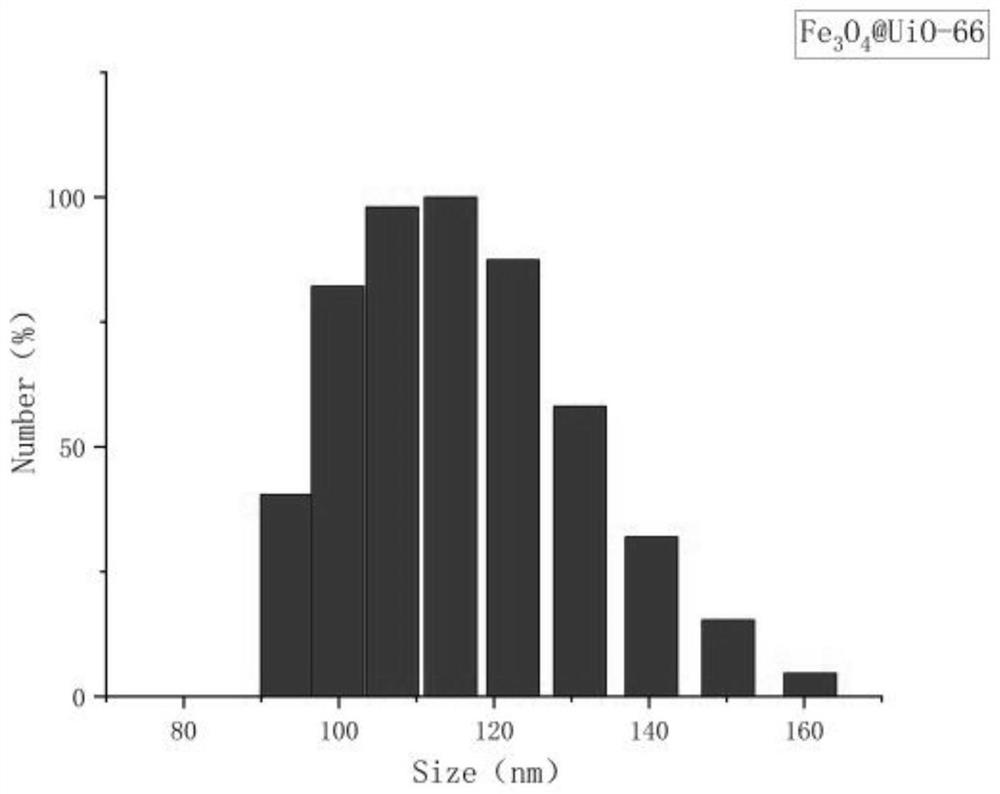
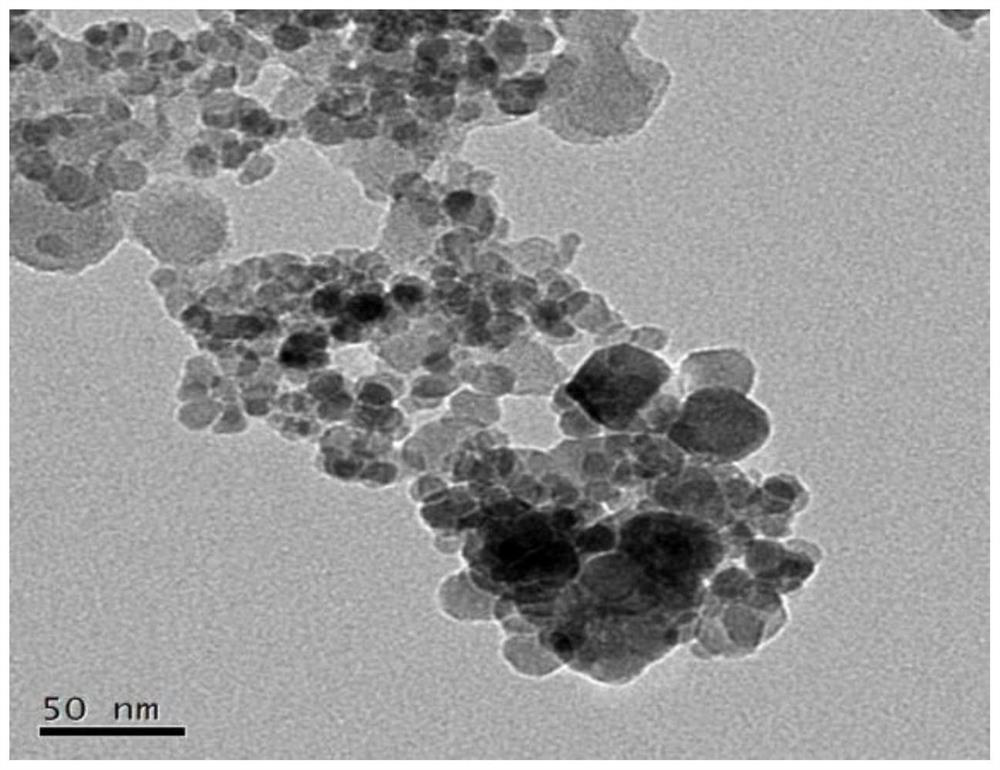
[0048] (1) Preparation of magnetic beads: adopt the hot solvent method to prepare, specifically: take 0.2g of ferric oxide, 0.15g of zirconium tetrachloride and 0.13g of terephthalic acid and dissolve them in 80mL of DMF, and then Add 4mL of acetic acid, sonicate for 10min to obtain a suspension; transfer the suspension to a 100mL polytetrafluoroethylene autoclave, then heat at 130°C for 12h, and cool to room temperature; finally, alternately wash with water and ethanol for 3 times, at 70°C Drying...
Embodiment 2
[0055] Use the kit in embodiment 1 to carry out the method for manually extracting bacterial genome DNA, concrete steps are as follows:
[0056] S1. Prepare LB liquid medium, inoculate activated strains, and culture at 200r / min at 37°C for 12-14h;
[0057] S2. Take 1ml of the mixed bacterial solution and centrifuge for 60s at a speed of 10000r / min;
[0058] S3, add 350 μL lysate and 20 μL proteinase K and incubate for 20 minutes;
[0059] S3. Add 350 μL of binding solution and 15 μL of magnetic bead suspension, mix well, let stand for 10 min, absorb the magnetic beads with a magnetic stand, and remove the supernatant;
[0060] S4. Add 700 μL of the first washing liquid, oscillate and mix well, and after 1 min, use a magnetic stand to absorb the magnetic beads and remove the supernatant; then add the second washing liquid 80% ethanol, mix well, and after 1 min, the magnetic stand will absorb the magnetic beads and remove the supernatant. clear solution, this step was repeated...
Embodiment 3
[0063] Use the kit in embodiment 1 to carry out the method for automatic extraction of bacterial genome DNA, concrete steps are as follows:
[0064] S1. Take 1ml of the mixture and add it to the EP tube, centrifuge at 10000r / min for 60s, and discard the supernatant;
[0065] S2. Add 350 μL lysate and 20 μL proteinase K and mix well for lysis and enzymatic hydrolysis;
[0066] S3. Add the mixed liquid to column 1 and column 7 of the 96 plate, and add 350 μL of binding solution (i.e. isopropanol, after adding, the concentration of isopropanol is 38%);
[0067] S4. Add 15 μL of magnetic bead suspension and 400 μL of deionized water to columns 2 and 8 of the 96 plate; add 600 μL of the first washing solution to columns 3 and 9 of the 96 plate, and add the second washing solution to columns 4 and 5 and columns 10 and 11; add 80 μL of eluate to columns 6 and 12 of the 96 plate;
[0068] S5. Put the 96 plate in the nucleic acid extractor, set the cleavage binding time to 8 minutes,...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com