DIP rapid amplification detection reagent for fully-integrated micro-fluidic chip and application of DIP rapid amplification detection reagent
A technology for detecting people and sequences, applied in the determination/testing of microorganisms, recombinant DNA technology, DNA/RNA fragments, etc., can solve problems such as population inference systems that have not been reported yet, and achieve convenient storage and transportation, simplified transportation methods, and high performance stable effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0088] Embodiment 1, the design of primers and the construction of AIDIP38 chip amplification system
[0089] 1. Primer design and synthesis
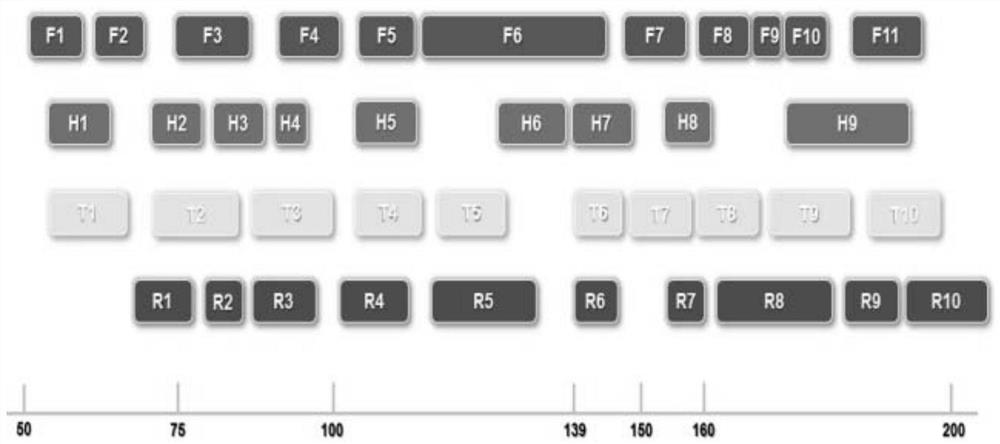
[0090] The present invention refers to related literature reports and dbSNP library, and obtains 38 InDel sites through screening (the information of each site is shown in Table 1). Primer premier 5.0 was used to design and optimize primer sequences based on the 38 InDel sites obtained from the above screening, as well as 1 sex site AMEL and 1 site DYS439 on the Y chromosome, and FAM, HEX, FRET-TAMRA and FRET- ROX is fluorescein-labeled at the 5' end of forward primer F at each site. Finally, the primer sequences designed for each locus were obtained, and all primer sequences were synthesized and labeled by Suzhou Xinhai Biotechnology Co., Ltd. See Table 2 for specific information on primer sequences and fluorescent markers, and see figure 1 .
[0091] Table 1. Information of each site
[0092]
[0093]
[0094] Table 2. Prim...
Embodiment 2
[0133] Embodiment 2, the application of AIDIP38 amplification reagent
[0134] 1. Experimental materials and methods
[0135] 1. Experimental materials
[0136] 1) Experimental samples
[0137] Volunteers provided 2 blood card samples, 2 saliva card samples, and 4 oral swab samples. The specific preparation method is as follows:
[0138] Oral swab samples: Scrape the left and right buccal mucosa of volunteers 8 times with the swab.
[0139] Blood card sample: Take 2 drops of blood from the fingertips of volunteers, each drop is about 1cm in diameter, and drop it in the marked circle of the blood collection card.
[0140] Saliva card sample: Take the mucous membrane swabs of the left and right cheeks of the volunteers and transfer them to the saliva card. The above samples are collected and dried in the shade for later use.
[0141] DNA standard 9947A (initial concentration: 10 ng / μL) is a product of Thermo Fisher Scientific, USA.
[0142] 2) Experimental equipment
[01...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com