Method for evaluating organic matter degradation potential of biological filter material
A bio-filter and microbial degradation technology, which is applied in the field of evaluating the potential of bio-filter to degrade organic matter, can solve the problems of long evaluation period, easy pollution, inconvenient operation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
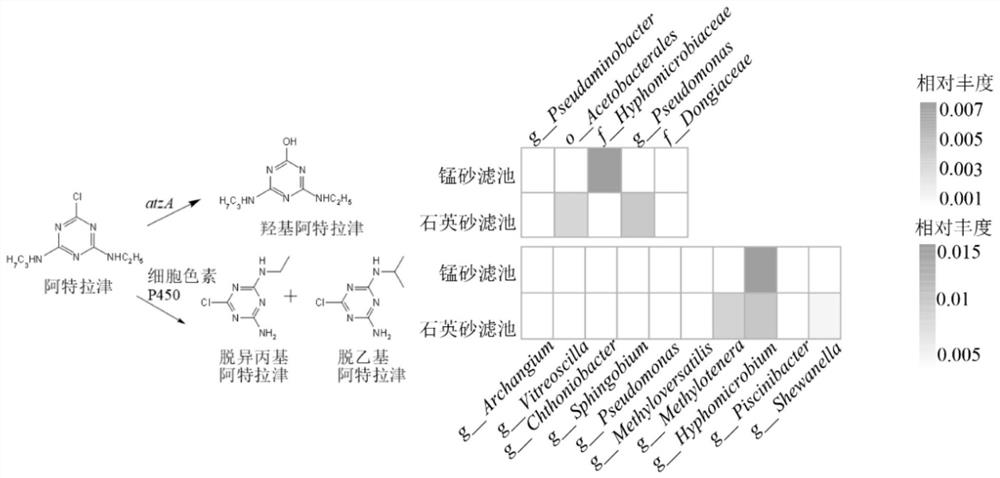
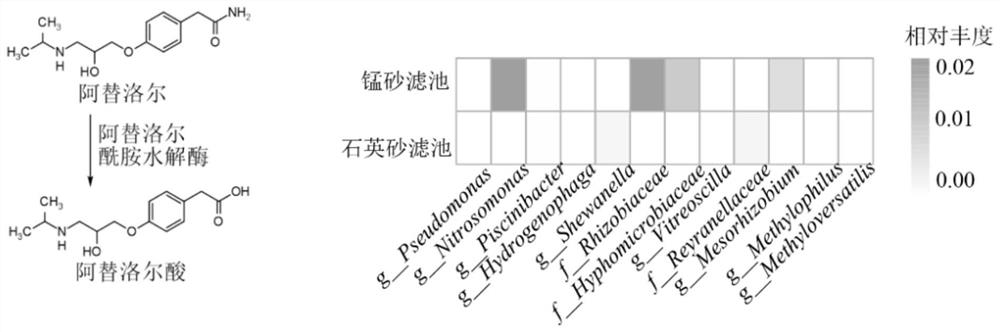
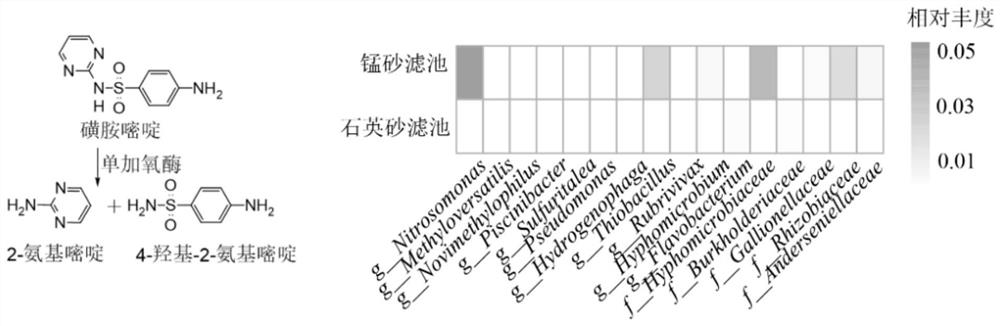
[0048] This embodiment provides a method for evaluating the potential of biological filter material to degrade organic matter. The target organic matter evaluated is atrazine, atenolol, sulfadiazine and carbamazepine respectively. The steps are as follows:
[0049] (1) Establish a reference database: determine the degradation genes and / or degradative enzymes (as shown in Table 1) of the microorganisms to the target organic matter according to the literature and the enviPath database; if the degradation genes can be found, obtain them in the Genbank database and / or the KEGG database For the DNA sequence of the degrading gene, if the degrading enzyme can be found, the protein sequence of the degrading enzyme is obtained in the Swiss-Prot database.
[0050] Table 1 Degradation genes, degrading enzymes and degradation pathways of target organic substances
[0051]
[0052]
[0053] The references in Table 1 are as follows:
[0054] [1]. De Souza, M.L.; Sadowsky, M.J.; Wacke...
Embodiment 2
[0081] The present embodiment provides a method to verify the reliability of the method for evaluating the potential of biological filter material to degrade organic matter in Example 1, the steps are as follows:
[0082] (1) Preparation of organic solution
[0083] Atenolol (Atenolol, C 14 h 22 N 2 o 3 ) was purchased from Shanghai Aladdin Biochemical Technology Co., Ltd.; Atrazine (Atrazine, C 8 h 14 ClN 5 ), sulfadiazine (Sulfadiazine, C 10 h 10 N 4 o 2 S) and carbamazepine (Carbamazepine, C 15 h 12 N 2 O) purchased from Shanghai McLean Biochemical Technology Co., Ltd.; Atenolol acid (Atenololacid, C 14 h 21 NO 4 ), Hydroxyatrazine (Hydroxyatrazine, C 8 h 15 N 5 O), deethylatrazine (Deethylatrazine, C 6 h 10 ClN 5 ), deisopropyl atrazine (Deisopropylatrazine, C 5 h 8 ClN 5 ), Carbamazepine 10,11-epoxide (Carbamazepine-10,11-epoxide, C 15 h 12 N 2 o 2 ), iminostilbene (Iminostilbene, C 14 h 11 N), 2-aminopyrimidine (2-Aminopyrimidine, C 4 h 5 ...
Embodiment 3
[0099] In this embodiment, the abundance of degrading genes and / or degrading enzymes is used to evaluate the potential of the biological filter material to degrade organic matter. The steps are as follows:
[0100] (1) Sampling
[0101] Sampling was carried out in quick sand filter pools of 9 drinking water plants nationwide, and the sampling method was referred to in Example 1.
[0102] (2) DNA extraction and metagenomic sequencing
[0103] Refer to Example 1 for the methods of DNA extraction and metagenomic sequencing.
[0104] (3) Calculate the abundance of degraded genes
[0105] Use Usearch v10.0.240 (-ublast-accel 0.8) to compare the DNA sequence in the metagenomic sequencing data obtained with the DNA sequence of the degraded gene in the reference database established in Example 1, to determine the microorganisms in the biofilter sample. Contains degradation genes;
[0106] Using the DNA sequence of the degraded gene as the reference sequence, add the length of the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com