Cytochrome P450 monooxygenase CYP109B2 mutant and application thereof
A CYP109B2, monooxygenase technology, applied in the field of biocatalytic enzymes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
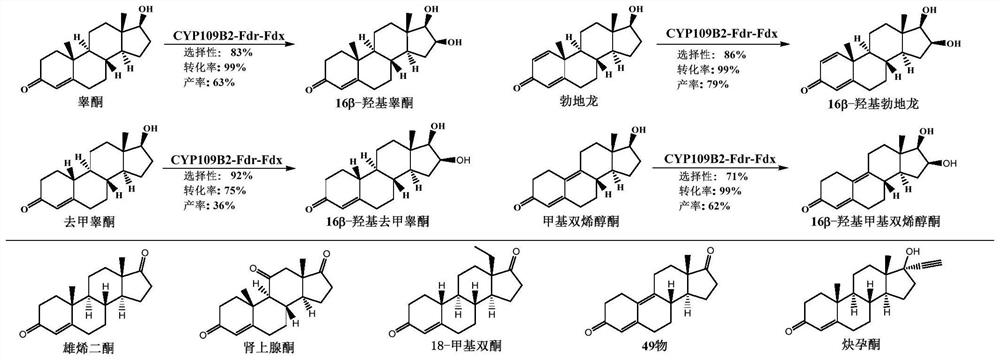
[0033] Example 1 Obtainment and Activity Verification of Cytochrome P450 Monooxygenase CYP109B2
[0034] 1. CYP109B2 gene cloning
[0035] Cultivate Sonora desert bacillus (Bacillus sonorensis) on seawater 2216 agar plate medium by conventional methods in the field, then extract the genome of Bacillus sonorensis by conventional methods in the field, and use this as a template to perform conventional PCR amplification to obtain the target Gene, the primers used are:
[0036] CYP109B2-F: ATGAACTCGGCAAAACAGCAGAAC (shown in SEQ ID NO:3)
[0037] CYP109B2-R: TCATGATGAAAGCAGCGCCTCTTTG (shown in SEQ ID NO:4)
[0038] The amplified product was detected by electrophoresis and sequenced. Its nucleotide sequence is shown in SEQ ID NO:2, and its translated amino acid sequence is shown in SEQ ID NO:1. Since this enzyme has 61.9% amino acid sequence homology with the reported CYP109 family CYP109B1, it was confirmed that the novel P450 monooxygenase is a member of the cytochrome monooxyg...
Embodiment 2
[0058] Construction of embodiment 2 CYP109B2 protease mutant library
[0059] 1. Determination of mutation sites
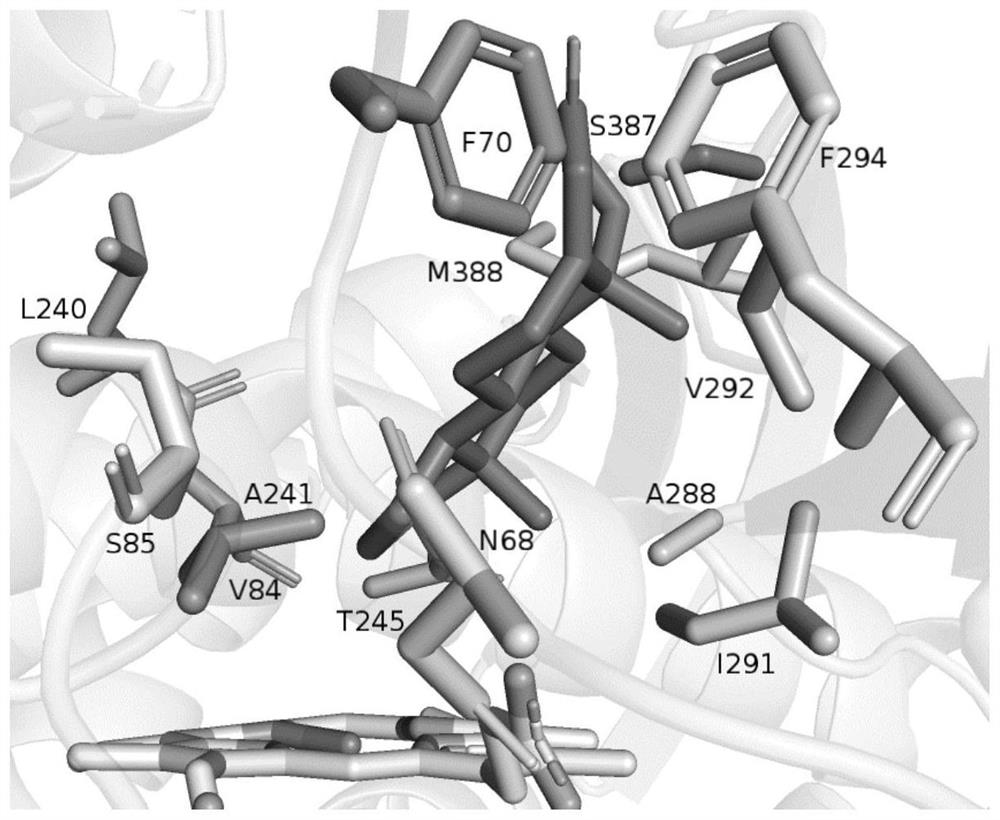
[0060] Using the homologous structure of CYP109B2 protein, the shape of the CYP109B2 active pocket and the binding form of the substrate testosterone to the enzyme molecular pocket were analyzed, and the molecular structure of the steroidal substrate testosterone was drawn by the scientific research drawing tool ChemBioDraw Ultra 14.0, and the protein crystal structure was used to analyze The software YASARA performed molecular dynamics simulation (MD) and molecular docking on the energy-minimized 3D molecular structure of testosterone and the obtained crystal structure of CYP109B2, and selected the molecules around the substrate testosterone The amino acid positions within the range are respectively N68, F70, V84, S85, L240, A241, T245, A288, I291, V292, F294, S387 and M388 (i.e. the 68th amino acid sequence shown in SEQ ID NO: 1 , 70, 84, 85, 240, 241, 245, 28...
Embodiment 3
[0069] Expression and enzyme activity identification of embodiment 3CYP109B2 mutant
[0070] 1. Expression of CYP109B2 mutant
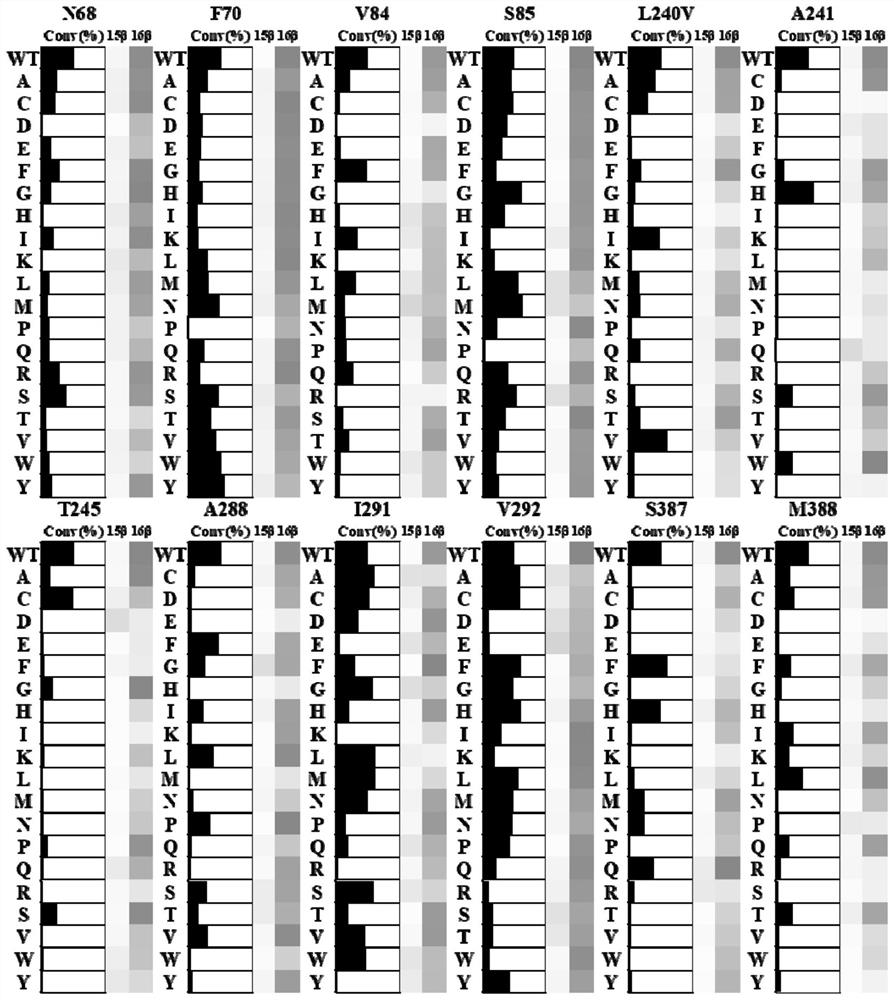
[0071] The PCR product obtained from the single-point saturation mutation and degenerate primer mutation in Example 2 was digested with the restriction endonuclease Dpn I, and the PCR product was transformed into Escherichia coli BL21 using a conventional transformation method in the art and After sequencing, all 19 amino acid mutants except the wild-type amino acid at the target site were obtained, indicating that the complete library of amino acid mutants at this site was completed. Then, the mutated nucleotide sequence was expressed according to the construction method of the recombinant cell E.coli (pRSFDuet-1-CYP109B2-Fdr_0978_Fdx_1499) in Example 2 to express the CYP109B2 mutant.
[0072] 2. Determination of catalytic activity and selectivity
[0073] After using the above method to express different CYP109B2 mutants, according to the detectio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com