Primer and probe for detecting delta 69/70 HV deletion mutation site of S gene of new coronavirus Alpha strain and application of primer and probe
A technology of deletion mutation and virus, applied in the field of biomedicine, can solve the problems of high cost and long time consumption, and achieve the effect of high specificity and sensitive signal
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1. Primers and probes for the S gene A69 / 70HV deletion mutation site of the new coronavirus Alpha strain
[0029] Due to the deletion of amino acids 69 and 70 in the S gene of the new coronavirus Alpha strain, if primers and probes designed based on the S gene of the original new coronavirus strain are used to detect the Alpha strain by RT-qPCR, it may not be detected New crown Alpha strain. After whole-genome sequencing verification, the S-gene target failure genotype (S-gene target failure, SGTF) was identified as a reliable marker for the Alpha strain. Select the S gene Δ69 / 70HV deletion site sequence of the Alpha strain to design primers and probes, which can be used as one of the effective proofs for determining the Alpha strain.
[0030] Table 1. Primers and probe sequences used in the present invention
[0031] sequence name Sequence information (5'-3') SEQ.ID.No Alpha strain S gene forward primer CTTGGTTCCATGCTATCTC 1 Alpha s...
Embodiment 2
[0033] Example 2. Detection of the S gene Δ69 / 70HV deletion mutation site of the new coronavirus Alpha strain
[0034] 1. Reagents required for detection
[0035] 1) Reagent I (1mL / tube): fluorescent quantitative PCR reaction buffer.
[0036] 2) Reagent II (lyophilized): a mixture of primers and probes for both the Alpha strain and the original strain (the concentration of the primer pair is 0.4 μmol / L, and the concentration of the probe is 0.2 μmol / L).
[0037] 3) Reagent III: Premix EX TaqTM enzyme mixture (TAKARA company).
[0038] 4) Positive quality control: including the synthetic new coronavirus Alpha strain S gene target fragment and including the new coronavirus original strain S gene target fragment.
[0039] 5) Negative quality control: water treated with diethyl pyrophosphate.
[0040] 6) Specimen types: nasal swab, throat swab, alveolar lavage fluid.
[0041] 2. Nucleic acid extraction
[0042]Use a commercial RNA extraction kit, such as a silica membrane spi...
Embodiment 3
[0055] Embodiment 3. Specific detection of primers and probes
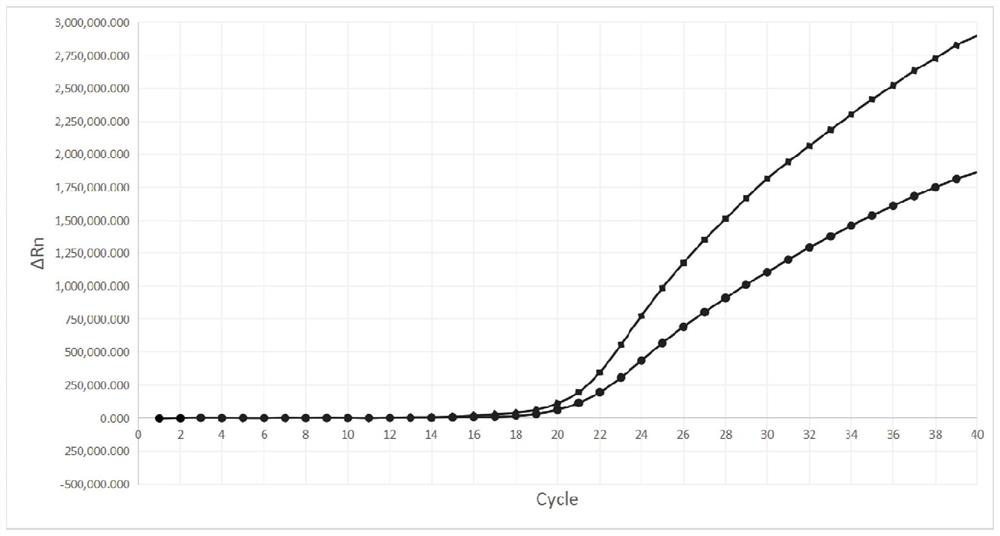
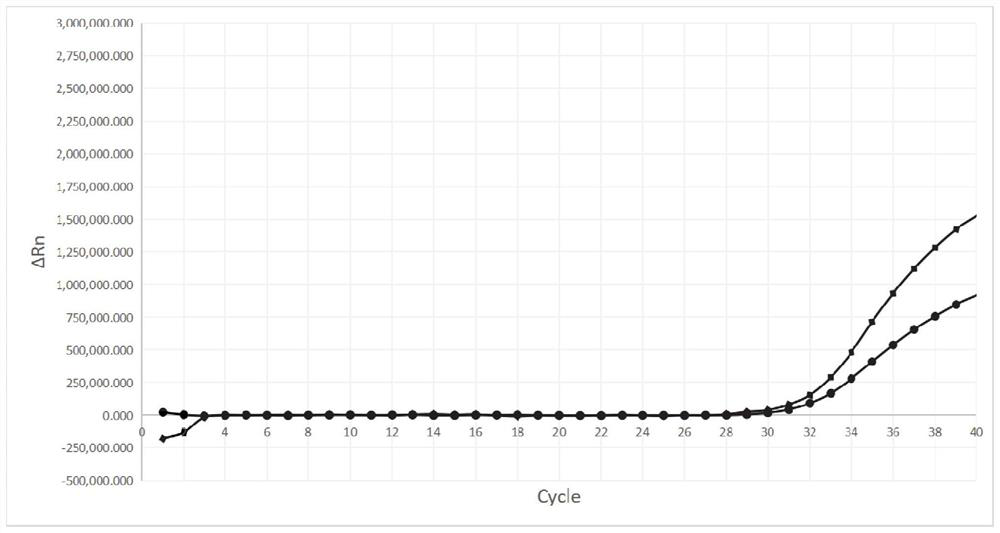
[0056] Add RNA fragments containing the full-length S gene of the new coronavirus Alpha strain, the original new coronavirus strain, SARS and MERS coronavirus respectively to the reaction tube, and detect according to the steps in Example 2, such as figure 1 and figure 2 As shown, the results show that amplification curve signals appeared in the FAM and HEX fluorescence channels respectively in the reaction tubes added with the Alpha strain of the new coronavirus and the original strain of the new coronavirus. In the reaction tubes added with RNA fragments of the full-length S gene of SARS and MERS coronaviruses, the Ct values of both FAM and HEX channels were greater than 30 (shown as negative), indicating that the designed Alpha strain of the new coronavirus, the original virus of the new coronavirus The strain primers and fluorescent probes have high reaction specificity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com