Top-speed RNA library building method and kit
A kit and extremely fast technology, applied in the biological field, can solve the problems of low detection rate of low-copy pathogens, low success rate of library construction, and low concentration, and achieve the effect of simple operation, wide application, and short process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: A method for extremely fast RNA library construction
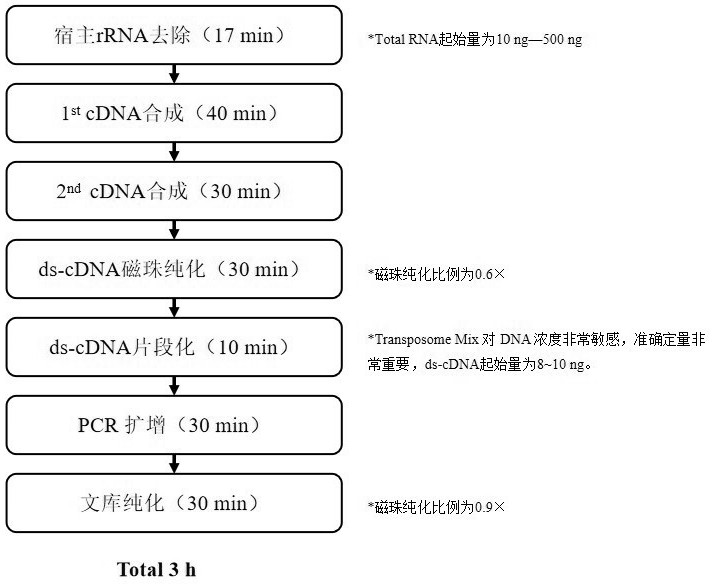
[0033] In this example, using the total RNA of 293 Cells, an extremely fast RNA library construction process was established. See the schematic diagram for the process and principle. figure 1 . The specific method is as follows:
[0034] 1) Host rRNA removal:
[0035] The Random primer used is NNNNNN (synthetic), 5.8S / 18S / 28S rRNA probe sequence reference CN202110257924 .X).
[0036] Table 1
[0037] components Dosage 293 Total RNA 10~500ng 4 μM 5.8S / 18S / 28S rRNA probe mix (CN202110257924 .X) 1 μL 1 mM Random Primer 1 μL Add DEPC water to 15 μL
[0038] 5 min at 85°C, 2 min at 72°C, 10 min at 60°C, and store at 4°C.
[0039] 2) Synthesis of 1st cDNA:
[0040] 1st Reaction buffer: 200 mM Tris-HCl, 250 mM KCl, 10 mM MgCl2, 5 mM dNTP, 20 mM DTT, pH 8.3.
[0041] Table 2
[0042] components Dosage The above reaction system 15μL 1st Re...
Embodiment 2
[0061] Example 2: Comparative analysis of an extremely fast RNA library construction method and traditional library construction methods
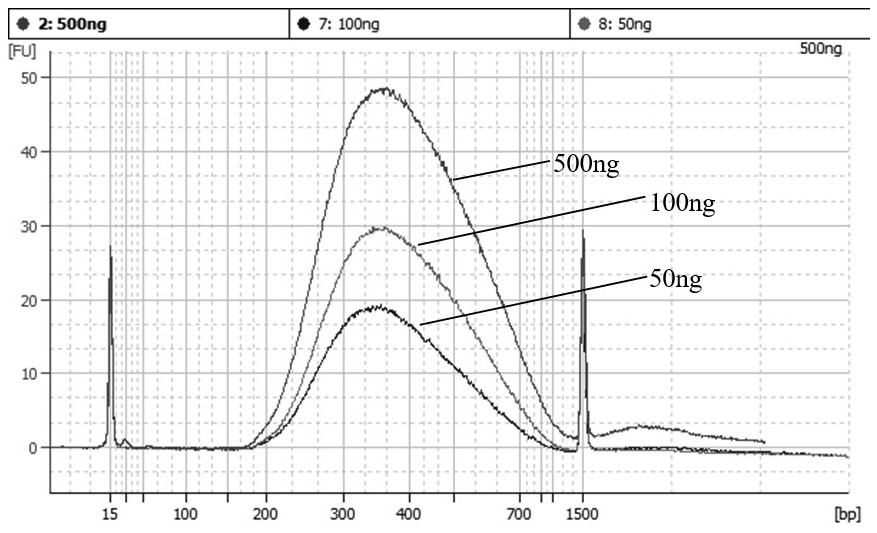
[0062] In this example, we compared the extremely fast RNA library construction method of the present invention with the traditional RNA library construction method. The traditional RNA library construction method uses NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina for library construction, the input amount of RNA is 500 ng, and the library construction process is carried out according to the product manual. In this extremely fast RNA library construction method, we have carried out library construction with RNA inputs of 500ng, 100ng, and 50ng, and the library construction process refers to Example 1.
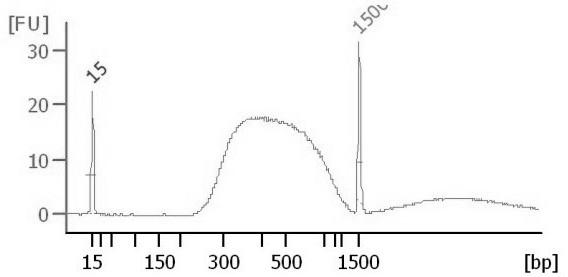
[0063] Qubit was used to quantify the constructed library, and Agilent 2100 Bioanalyzer was used to verify the library size distribution. The constructed library was sequenced and analyzed on the Illumina NovaSeq 6000 pla...
Embodiment 3
[0067] Example 3: Application test of a kit for rapid RNA library construction in pseudovirus detection
[0068] In this example, we will COVID-19-pseudovirus (2019-nCOV pseudovirus) (Yeasen, 11900) according to 10 4 -10 7 Add 2.5 μL to 50 ng of 293 Total RNA at the ratio of cells / mL, use an extremely fast RNA library building kit to build a library and send it for sequencing.
[0069] COVID-19-pseudovirus (2019-nCOV pseudovirus) (Yeasen, 11900) retroviral vector, the vector is loaded with the partial sequence of the COVID-19 virus ORF1 a / b gene, the entire sequence of the E gene and the N gene coding region, where N The gene sequence is shown in SEQ ID No.1. According to the N gene sequence, we used QRT-PCR to detect the primers ORF1ab-F: 5´-GTGARATGGTCATGTGTGGCGG-3´, ORF1ab-R: 5´-CARATGTTAAASACACTATTAGCATA-3´, and detected the RNA of COVID-19-pseudovirus after 1st cDNA synthesis 1st cDNA synthesis efficiency. Quantitative PCR analysis was performed on the 100-fold dilute...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com