KASP molecular marker for high-throughput detection of high oleic acid mutation site of cotton GhFAD2-1 gene
A technology of molecular markers and mutation sites, which is applied in the direction of DNA/RNA fragments, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of long detection cycle, low detection efficiency, high detection cost, etc., and achieve long detection cycle, Low throughput, high cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: Development of closely linked KASP markers for cotton high oleic trait genes GhFAD2-1A and GhFAD2-1D
[0029] 1. KASP primer design
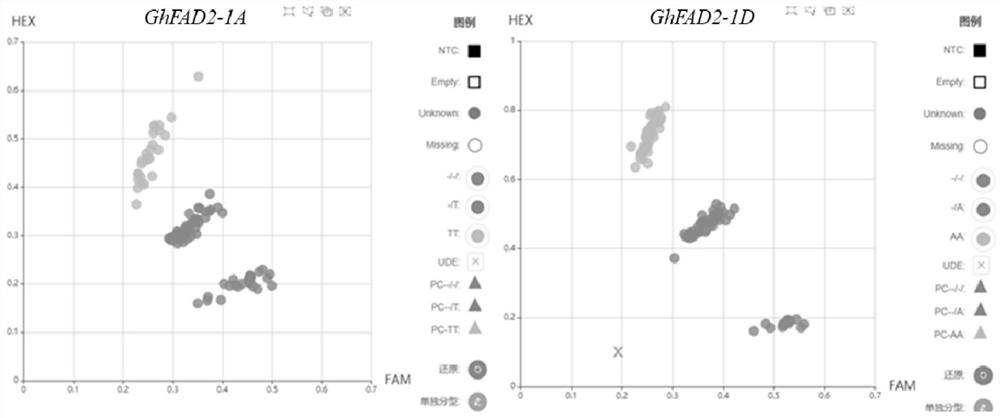
[0030] Extract the 100 bp flanking sequences around the 429nt site of the GhFAD2-1A / D gene coding region, use Primer5.0 software to design 3 sets of KASP primers, and use the ArrayTape platform of Douglas Scientific Company to detect the amplification effects of KASP-A429 and KASP-D429 And the polymorphism is the best, and the InDel difference between GhFAD2-1A and GhFAD2-1D in wild type and high oleic acid mutant can be clearly distinguished.
[0031] The primers for the molecular marker KASP-A429 are:
[0032] 1) Two specific primers:
[0033] GhFAD2-1A-HEX: 5'-GAAAATCAGTCACCGCCGTCAt-3', as shown in SEQ ID No.3;
[0034] GhFAD2-1A-R: 5'-ACTCGACCGGGTGGATTGTTT-3', as shown in SEQ ID No.4;
[0035] 2) A universal primer:
[0036] GhFAD2-1AD-FAM: 5'-GAAAATCAGTCACCGCCGTCAC-3', as shown in SEQ ID No.5;
[0037] The primers fo...
Embodiment 2
[0054] Example 2: Application of KASP markers
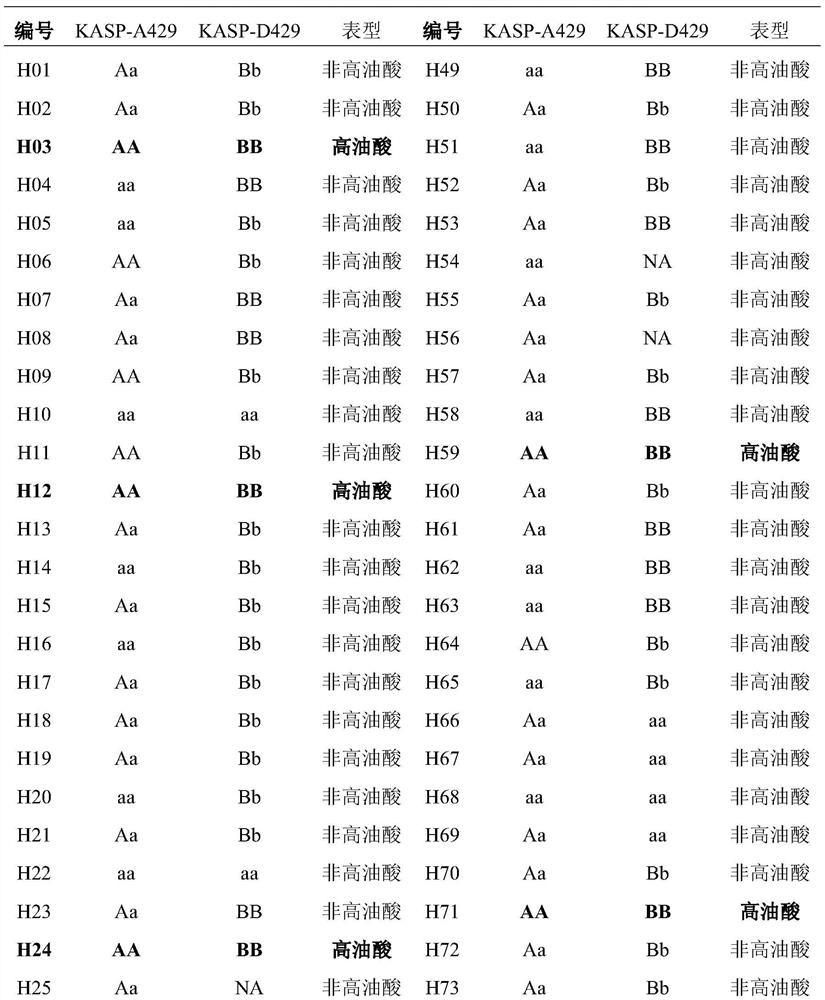
[0055] In order to verify the practicability of the KASP marker of the present invention, the cotton materials described in Table 1 were used for detection and analysis. Among them, H01-H92 is Lumian 418 as the female parent and high oleic acid cotton mutant m1-2 (Chen Yizhen, Fu Mingchuan, LiHao, et al., High-oleic acid content, nontransgenic allotetraploid cotton (Gossypium hirsutum L.) generated by knockout of GhFAD2 genes with CRISPR / Cas9system.Plant Biotechnology Journal, 2021,19:424-426.) The F2 segregation population prepared for male parent crosses, H93 and H94 are high oleic acid cotton mutant m1-2, used as Positive control, H95 and H96 are Lumian 418, used as negative control.
[0056] Table 1 Phenotype and genotype information of cotton segregation population KASP primer list
[0057]
[0058]
[0059] The results are shown in Table 1 and figure 1 As shown, after KASP analysis, among the 92 individual plants o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com