Enteric microorganism combination and application thereof as systemic lupus erythematosus marker
A technology for gut microbes and lupus erythematosus, applied in the field of microbiology, can solve problems such as lack of early detection methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Example 1 Study Subjects and Sample Collection
[0075] With informed consent, fecal samples from 300 subjects, including 120 SLE patients and 180 healthy controls, were collected at the Nanfang Hospital of Southern Medical University.
[0076] To be eligible for inclusion in this study, individuals must meet the following criteria for stool sample collection:
[0077] 1) No treatment with antibiotics or immunomodulators, no specific diet (diabetics, vegetarians, etc.) and normal lifestyle (no extra stress) for at least 3 months;
[0078] 2) At least 3 months after any medical intervention;
[0079] 3) No pregnancy, tumor, infection or autoimmune disease or rheumatic disease
[0080] Distribute stool sample collection tubes to the research subjects and inform them of the specific collection method. The subjects take 10-15g of fresh feces and put them in a collection tube with fecal genome protection solution, express them to the hospital laboratory at room temperature...
Embodiment 216
[0081] Example 2 16S rRNA Analysis of Intestinal Microflora
[0082] 2.1 DNA extraction
[0083] DNA extraction was performed using a Fecal Microbial Genome Extraction Kit (Guangdong Nanxin Medical Technology Co., Ltd.) according to the manufacturer's instructions.
[0084] Extracts were treated with DNase-free RNase to eliminate RNA contamination. DNA content was determined using a NanoDrop spectrophotometer, Qubit fluorometer.
[0085] 2.2 DNA library construction and sequencing
[0086] The purified genomic DNA was subjected to two rounds of 16S rRNA gene PCR amplification with bacterial universal primers to complete the construction of the sequencing sample library. 2.0 Fluorometer (Invitrogen, USA) detection concentration, Agilent2100 (Agilent, USA) detection library size; Application Illumina MiSeq (Illumina, San Diego, USA) sequencer is used to sequence the sequence of the amplified 16S rRNA hypervariable region, and the sequencing region is V3-V4 area.
[0087] 2....
Embodiment 3
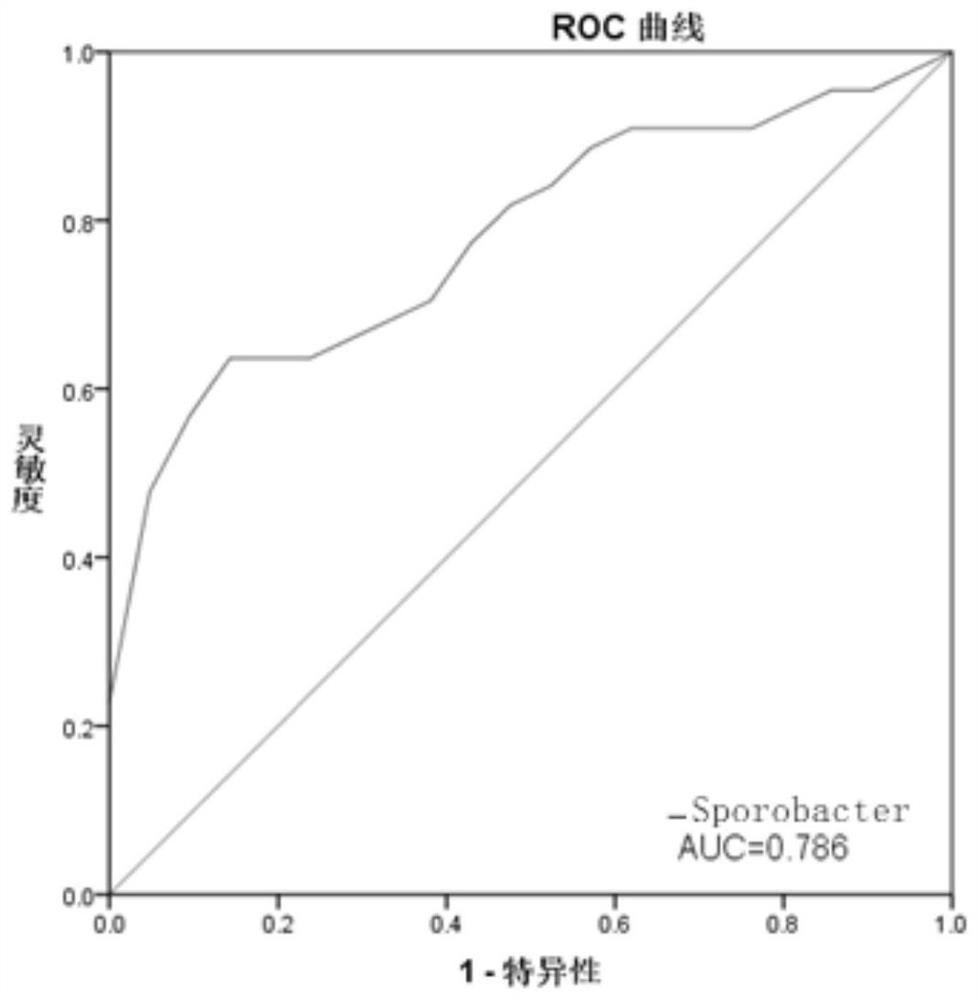
[0097] Example 3 Clinical detection and verification of intestinal flora in SLE patients
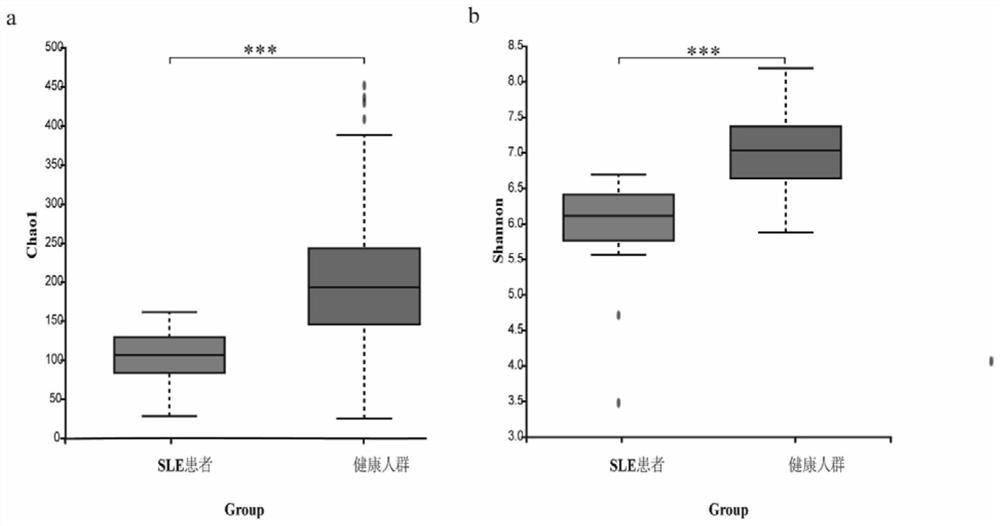
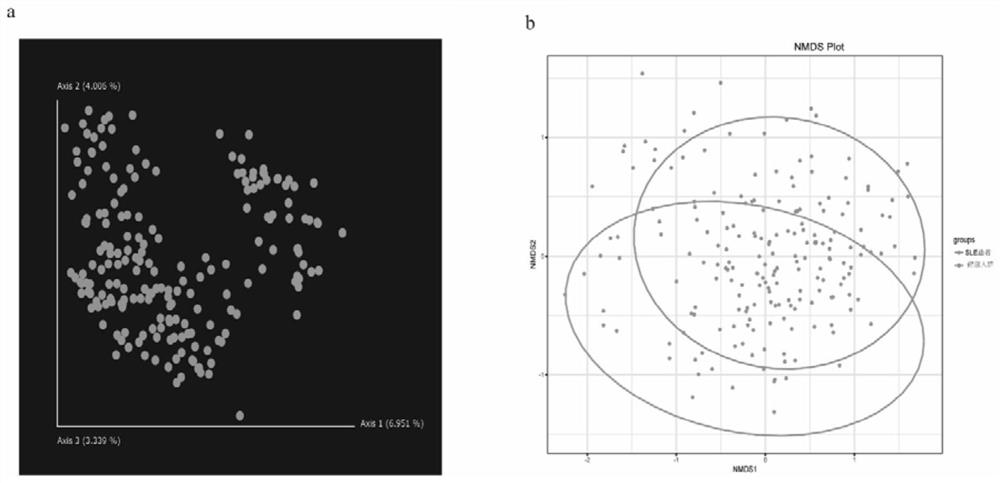
[0098] The above studies have shown that SLE patients are significantly different from healthy people in the classification of intestinal bacteria species, and there are differences in the level of multiple intestinal microorganisms. ).
[0099] According to the correlation between related flora and SLE, SLE can be diagnosed by detecting the abundance of related flora in patient samples.
[0100] Therefore, in this embodiment, 100 feces samples from healthy patients and 100 SLE patients were collected, and the samples were numbered for testing. According to the 16S rRNA gene sequence of the target microorganism, specific fluorescent quantitative PCR primer probes were designed and internal standards were set.
[0101] Table 2 shows the real-time fluorescent quantitative PCR (Quantitative Real-time PCR) specific primer sequences for detecting the abundance of Sporobacter, Butyricimonas,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com