Escherichia coli genetic engineering strain for exocytosis expression of Ulp1 protease and application of escherichia coli genetic engineering strain
A technology of genetically engineered strains and Escherichia coli, applied in the field of protein expression, can solve the problems of slow growth rate, low secretion and expression yield, and expensive medium, and achieve the effects of low cost, high cutting activity and high cutting efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
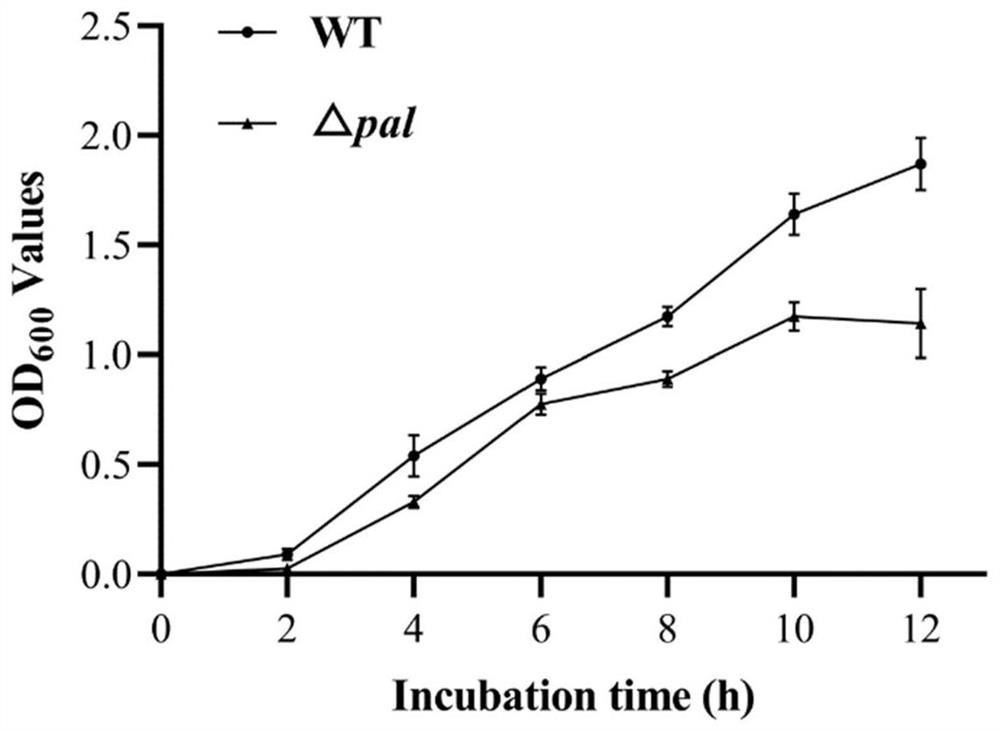
[0052] Construction of the Escherichia coli BL21 (DE3) bacterial strain of embodiment 1 pal gene deletion
[0053] The gene knockout of Escherichia coli BL21(DE3) uses the Red homologous recombination technology, and the plasmids used include pKD46, pKD13 and pCP20.
[0054] 1. Construction of Escherichia coli BL21(DE3)△pal:kan strain
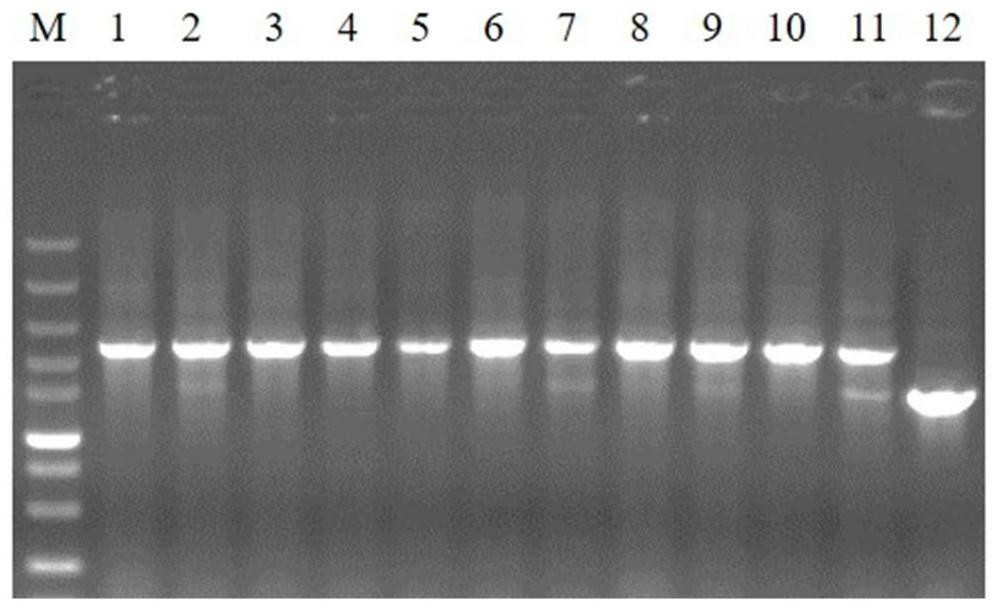
[0055] (1) According to the upstream and downstream sequences of the pal gene in the Escherichia coli BL21 (DE3) genome published by NCBI, two pairs of primers were designed using pKD13 as a template: 5′pal-FRT-1 / 3′pal-FRT-1 and 5′pal- FRT-2 / 3'pal-FRT-2.
[0056] (2) Use pKD13 as a template, 5'pal-FRT-1 and 3'pal-FRT-1 as primers for the first round of PCR; use the recovered product of the first round of PCR as a template, 5'pal-FRT-2 and 3'pal-FRT-2 and 3' 'pal-FRT-2 is used as a primer for the second round of PCR. Through these two rounds of PCR reactions, a 70bp homology arm sequence at both ends and a PCR product containing a kanamycin re...
Embodiment 2
[0073] Example 2 Escherichia coli BL21 (DE3) △ pal strain extracellular secreted method for expressing Ulp1 protease
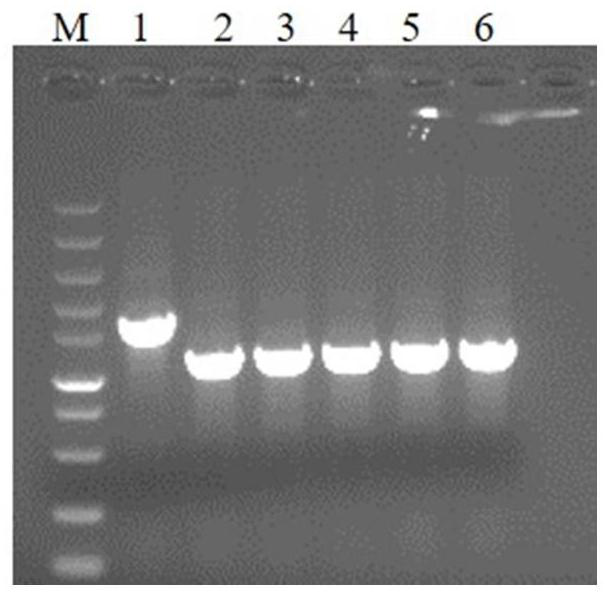
[0074] 1. Construction of extracellular secreted expression Ulp1 vector and strain
[0075] According to the nucleotide sequence of the 403-621 amino acid region of the Ulp1 gene published by NCBI, the nucleotide sequence encoding the SX signal peptide, the nucleotide sequence encoding the Linker whose amino acid sequence is RGSGG, and 6 histidine residues were introduced into its N-terminus. Base sequence, fusion gene structure such as Figure 4 As shown, the size of the fusion gene is 747bp, and the size of the fusion protein is 28.7kDa. The fusion gene was cloned into the downstream of the RBS of pET22b with NdeI and XhoI, named pET22b-SX-6His-Linker-Ulp1, see the vector structure Figure 5 . The expression vector pET22b-SX-6His-Linker-Ulp1 was transformed into Escherichia coli BL21(DE3)△pal strain to obtain the Escherichia coli strain expressing Ulp1 by...
Embodiment 3
[0080] Example 3 Detection of the expression of Ulp1 protease secreted extracellularly
[0081] Whether the Ulp1 protease expressed extracellularly in Example 2 is expressed or not is determined by SDS-PAGE. The extracellular secreted Ulp1 protease contains a 6×His tag, and Ni NTAbeads can be added to the expression supernatant for specific enrichment and concentration, which is helpful for protein detection. Specific steps are as follows:
[0082] (1) Take 4ml of the bacterial liquid after induced expression and centrifuge at 13000rpm for 10min at 4°C.
[0083] (2) Collect the supernatant after centrifugation, filter it with a 0.45 μm filter membrane, add 20 μl Ni NTAbeads, and rotate at 4° C. for 1 h.
[0084] (3) After the supernatant is fully combined with the beads, centrifuge at 3000 rpm for 5 min at 4°C and discard the supernatant. Add 100 μl of protein loading buffer to the remaining beads, cook at 95°C for 5 minutes.
[0085] (4) SDS-PAGE electrophoresis was perfo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com