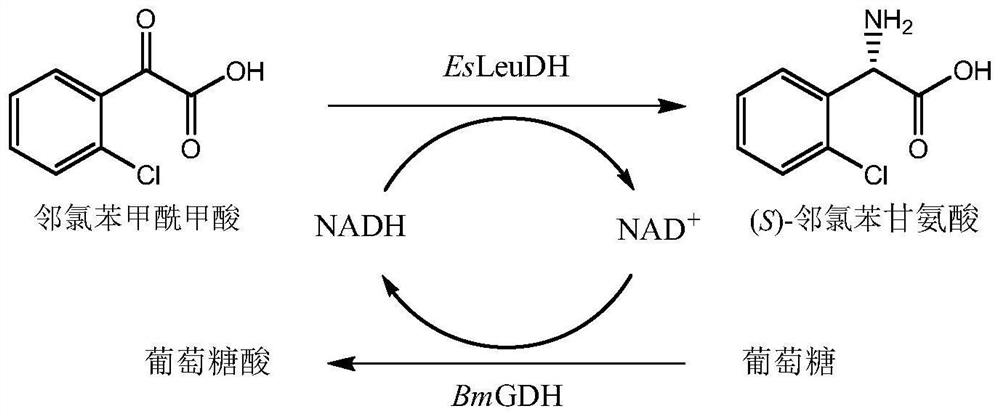
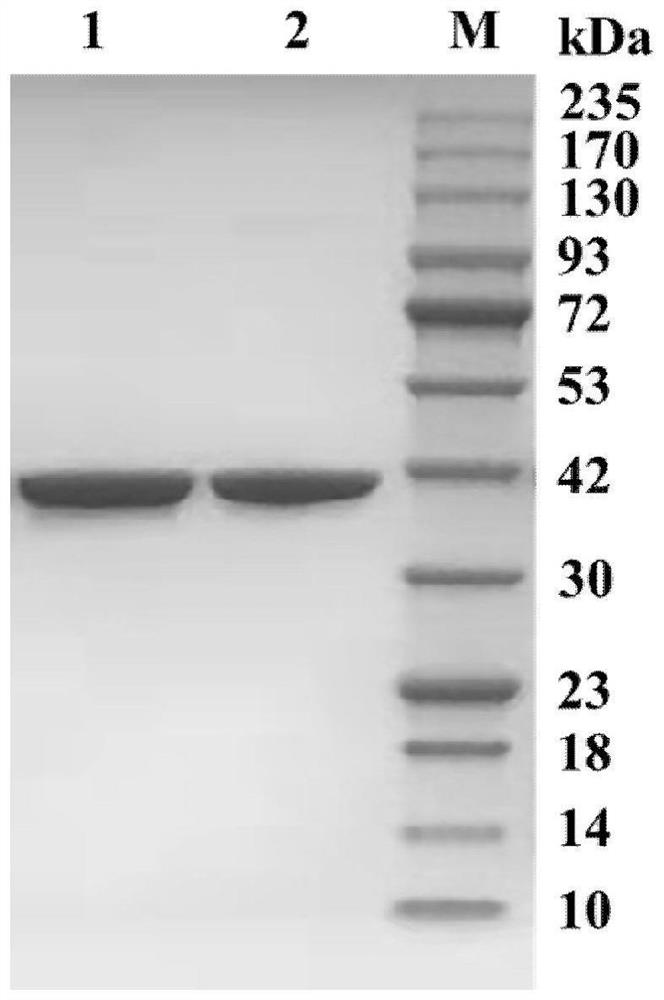
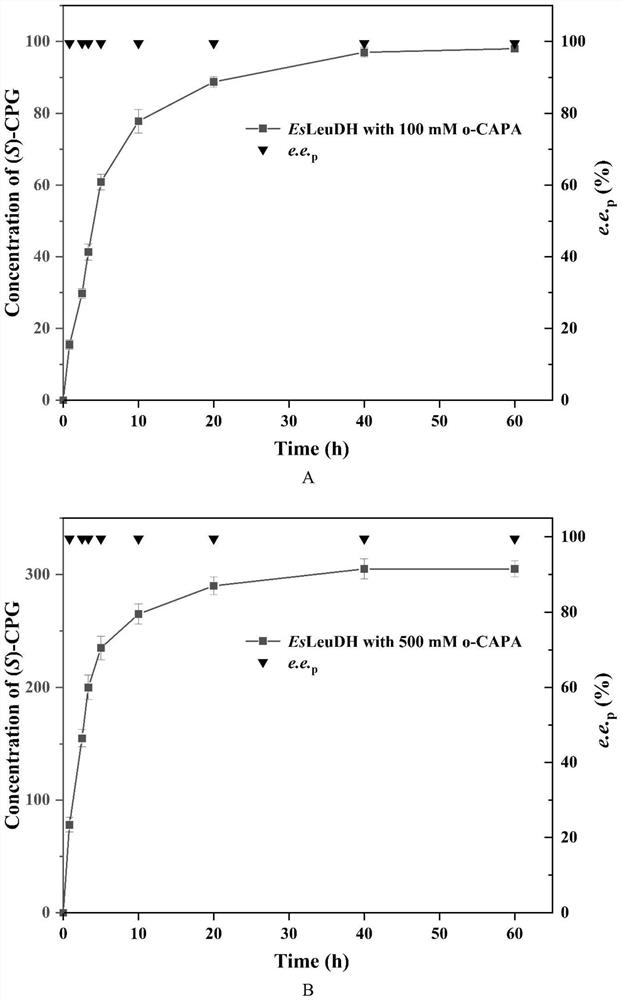
Leucine dehydrogenase mutant and application thereof in synthesis of (S)-2-chlorophenylglycine
A technology of leucine dehydrogenase and o-chlorophenylglycine, which is applied to leucine dehydrogenase mutant and its application field in the synthesis of (S)-o-chlorophenylglycine, and can solve the problem of low substrate feeding amount , the problem of low asymmetric amination activity of o-chlorobenzoylformic acid, etc., to achieve the effect of high catalytic rate, high product e.e value and mild reaction conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: Construction and screening of leucine dehydrogenase random mutation library
[0029] The vector pET28a(+)-esleudh (leucine dehydrogenase esleudh amino acid sequence shown in SEQ ID NO.2 in the starting strain E.coli BL21(DE3) / pET28a(+)-esleudh, encoding gene nucleotide sequence As shown in SEQ ID NO.1) as a template, using epPCR-F and epPCR-R in Table 1 as primers, using an error-prone PCR kit (purchased from Tianenze Company, ready-to-use error-prone PCR kit) to construct random mutations library.
[0030] EsLeuDH nucleotide sequence:
[0031] atggtggaaacgaacgttgaggcgcgcttctcgattttcgagacgatggcgatggaagactacgaacaggtggttttttgccatgacaaagtgagcgggctgaaagcaattattgcgattcatgataccaccctgggtccggcactgggtggcctgagaatgtggaattatgcaagcgatgaagaagcattaattgatgcattacgcctggcaaaaggtatgacctacaaaaacgcagcagcaggcctgaatttaggtggtggtaaagcggttataatcggggacgccaaaacccagaaaagcgaagcactgtttagagcatttggtcgttacgttcagagcctgaacggaagatatattacagcagaggatgttaatacaacagttgcagatatggattatatccatatggaaacagatt...
Embodiment 2
[0039] Example 2: Inducible expression of wild-type leucine dehydrogenase, mutants and glucose dehydrogenase
[0040] Glucose dehydrogenase genetically engineered bacteria: according to the glucose dehydrogenase gene (Genbank accession number: 1RWB_A) from Bacillus megaterium (Bacillus megaterium) IWG3, artificially synthesized glucose dehydrogenase bmgdh gene (nucleotide sequence such as SEQ ID NO. 3, the amino acid sequence of the encoded protein (as shown in SEQ ID NO.4) was inserted between the Nco I and Xho I restriction sites of pET28b (+) to construct a recombinant expression vector, and this expression vector was transferred into E. coli BL21(DE3), E. coli BL21(DE3) / pET28b(+)-bmgdh was produced.
[0041] bmgdh nucleotide sequence:
[0042] ATGTACAAGGACCTTGAGGGAAAGGTCGTCGTCATTACTGGATCTTCTACTGGACTGGGAAAGTCTATGGCTATTCGATTCGCTACTGAGAAGGCTAAGGTCGTCGTGAACTACCGATCTAAGGAGGACGAGGCTAACTCTGTCCTTGAGGAGATTAAGAAGGTCGGAGGAGAGGCTATTGCTGTCAAGGGTGACGTCACTGTCGAGTCTGACGTCATTAACCTGGTCCAGT...
Embodiment 3
[0045] Embodiment 3: Enzyme activity assay of wild-type leucine dehydrogenase
[0046] 1. Determination of enzyme activity
[0047] The starting strain E.coli BL21 (DE3) / pET28a (+)-esleudh wet thallus and E.coli BL21 (DE3) / pET28b (+)-bmgdh wet thallus with dry weight ratio 2: 1(w / w) mixed into the starting mixed cells.
[0048] Enzyme activity unit (U) is defined as: under the conditions of 40°C and pH 9.0, the amount of enzyme required to produce 1 micromole of (S)-o-chlorophenylglycine per minute is defined as an enzyme activity unit, U. Specific enzyme activity is defined as the number of activity units per mg of enzyme protein, U / mg.
[0049] Enzyme activity assay method: use the starting mixed bacteria as a catalyst, o-chlorobenzoylformic acid as a substrate, glucose as an auxiliary substrate, and add a small amount of exogenous NAD + , with pH 9.0, 100mM Tris-HCl buffer as the reaction medium, a coenzyme circulation system was established. The reaction system was sel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com